Model-based analyses of whole-genome data reveal a complex evolutionary history involving archaic introgression in Central African Pygmies - PubMed (original) (raw)
Model-based analyses of whole-genome data reveal a complex evolutionary history involving archaic introgression in Central African Pygmies
PingHsun Hsieh et al. Genome Res. 2016 Mar.
Erratum in
- Corrigendum: Model-based analyses of whole-genome data reveal a complex evolutionary history involving archaic introgression in Central African Pygmies.
Hsieh P, Woerner AE, Wall JD, Lachance J, Tishkoff SA, Gutenkunst RN, Hammer MF. Hsieh P, et al. Genome Res. 2016 May;26(5):717.1. doi: 10.1101/gr.206524.116. Genome Res. 2016. PMID: 27197244 Free PMC article. No abstract available.
Abstract
Comparisons of whole-genome sequences from ancient and contemporary samples have pointed to several instances of archaic admixture through interbreeding between the ancestors of modern non-Africans and now extinct hominids such as Neanderthals and Denisovans. One implication of these findings is that some adaptive features in contemporary humans may have entered the population via gene flow with archaic forms in Eurasia. Within Africa, fossil evidence suggests that anatomically modern humans (AMH) and various archaic forms coexisted for much of the last 200,000 yr; however, the absence of ancient DNA in Africa has limited our ability to make a direct comparison between archaic and modern human genomes. Here, we use statistical inference based on high coverage whole-genome data (greater than 60×) from contemporary African Pygmy hunter-gatherers as an alternative means to study the evolutionary history of the genus Homo. Using whole-genome simulations that consider demographic histories that include both isolation and gene flow with neighboring farming populations, our inference method rejects the hypothesis that the ancestors of AMH were genetically isolated in Africa, thus providing the first whole genome-level evidence of African archaic admixture. Our inferences also suggest a complex human evolutionary history in Africa, which involves at least a single admixture event from an unknown archaic population into the ancestors of AMH, likely within the last 30,000 yr.
© 2016 Hsieh et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
Figure 1.
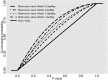
Significant excess of tests with very low _P_-values in the observed _S* P_-value distribution. Plotted is the observed (dashed lines) _S* P_-value distribution for the real data, calculated based on each of the four sets of whole-genome simulations. The four sets arise from the combination of the two demographic null models (Supplemental Fig. S2): Model-1 (the continuous asymmetric gene flow model) and Model-2 (the single-pulse admixture model); and the two genetic recombination maps: HapMap Yoruba map (HapMap) and African American map (AAMap). The solid line represents the null _S* P_-value distributions of the four whole-genome null simulation sets, derived by calculating _P_-values using a single randomly chosen simulation from each set. All four are uniformly distributed between 0 and 1 as expected. For the real data (dashed lines), all four analyses show a significant shift to small _P_-values in the observed _S* P_-value distribution (one-sided Mann-Whitney U test, P < 2.2 × 10−16), thus rejecting our demographic null hypotheses including no archaic admixture.
Figure 2.
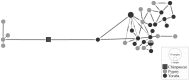
Haplotype network for the candidate introgressive locus Chr 16:8702222-8747116. Each circle is a haplotype with size proportional to the haplotype frequency, and the shade of gray indicates the haplotype frequency in the Pygmy (lighter gray) and Yoruba (darker gray) samples. Vertical bars along each branch indicate the number of mutations separating the haplotypes.
Figure 3.
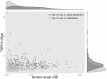
Wide joint distribution of TMRCA and genetic length for the top 1% _S* P_-value candidate loci. Darker gray dots and lighter gray triangles are, respectively, the two candidate sets from the top 1% candidate loci from the observed _S* P_-value and empirical S* distributions. The top and right plots show the marginal density of genetic length and TMRCA, respectively, for both candidate sets.
Figure 4.
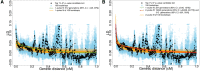
Decay of pairwise LD with respect to genetic distance for SNPs ascertained from the top 1% candidate introgressive loci. Black and blue dots are the average estimated LD among pairs of SNPs binned using genetic distance (in 0.001 cM increments) using real data and 100 bootstraps, respectively. The genetic distance is calculated based on the HapMap Yoruba map (The International HapMap Consortium 2007). For the cases of using the African American map (Hinch et al. 2011), see Supplemental Figure S10. The green curve is the fit of a single exponential using the data, while the red and orange curves are the fits of two exponentials using the real data and 100 bootstraps, respectively. (A) Fitting LD decay within genetic distance 0.02–1 cM. (B) Fitting LD decay within genetic distance 0.002–1 cM.
Figure 5.
Comparison of the joint distributions of TMRCA and genetic length for the top 1% _S* P_-value candidate loci from the data and whole-genome archaic admixture simulations. In each panel, the joint and marginal distributions of TMRCA (million yr ago) and genetic length (cM) of our candidate loci from the data (black cross and solid line in scatter and density plots, respectively) are compared with those from archaic admixture simulations (symbols and dashed lines in scatter and density plots, respectively): (A) two-wave archaic admixture model; (B) single-wave, 2% archaic admixture; (C) single-wave, 5% archaic admixture. TMRCA estimates for archaic simulation candidates were obtained from simulated coalescent trees in MaCS (Chen et al. 2009).
Similar articles
- Whole-genome sequence analyses of Western Central African Pygmy hunter-gatherers reveal a complex demographic history and identify candidate genes under positive natural selection.
Hsieh P, Veeramah KR, Lachance J, Tishkoff SA, Wall JD, Hammer MF, Gutenkunst RN. Hsieh P, et al. Genome Res. 2016 Mar;26(3):279-90. doi: 10.1101/gr.192971.115. Epub 2016 Feb 17. Genome Res. 2016. PMID: 26888263 Free PMC article. - Genetic evidence for archaic admixture in Africa.
Hammer MF, Woerner AE, Mendez FL, Watkins JC, Wall JD. Hammer MF, et al. Proc Natl Acad Sci U S A. 2011 Sep 13;108(37):15123-8. doi: 10.1073/pnas.1109300108. Epub 2011 Sep 6. Proc Natl Acad Sci U S A. 2011. PMID: 21896735 Free PMC article. - Whole-genome sequence analysis of a Pan African set of samples reveals archaic gene flow from an extinct basal population of modern humans into sub-Saharan populations.
Lorente-Galdos B, Lao O, Serra-Vidal G, Santpere G, Kuderna LFK, Arauna LR, Fadhlaoui-Zid K, Pimenoff VN, Soodyall H, Zalloua P, Marques-Bonet T, Comas D. Lorente-Galdos B, et al. Genome Biol. 2019 Apr 26;20(1):77. doi: 10.1186/s13059-019-1684-5. Genome Biol. 2019. PMID: 31023378 Free PMC article. - Archaic human genomics.
Disotell TR. Disotell TR. Am J Phys Anthropol. 2012;149 Suppl 55:24-39. doi: 10.1002/ajpa.22159. Epub 2012 Nov 2. Am J Phys Anthropol. 2012. PMID: 23124308 Review. - Searching for archaic contribution in Africa.
Santander C, Montinaro F, Capelli C. Santander C, et al. Ann Hum Biol. 2019 Mar;46(2):129-139. doi: 10.1080/03014460.2019.1624823. Epub 2019 Jun 26. Ann Hum Biol. 2019. PMID: 31163991 Review.
Cited by
- Evolutionary and Medical Consequences of Archaic Introgression into Modern Human Genomes.
Dolgova O, Lao O. Dolgova O, et al. Genes (Basel). 2018 Jul 18;9(7):358. doi: 10.3390/genes9070358. Genes (Basel). 2018. PMID: 30022013 Free PMC article. Review. - The influence of evolutionary history on human health and disease.
Benton ML, Abraham A, LaBella AL, Abbot P, Rokas A, Capra JA. Benton ML, et al. Nat Rev Genet. 2021 May;22(5):269-283. doi: 10.1038/s41576-020-00305-9. Epub 2021 Jan 6. Nat Rev Genet. 2021. PMID: 33408383 Free PMC article. Review. - Genetics and Material Culture Support Repeated Expansions into Paleolithic Eurasia from a Population Hub Out of Africa.
Vallini L, Marciani G, Aneli S, Bortolini E, Benazzi S, Pievani T, Pagani L. Vallini L, et al. Genome Biol Evol. 2022 Apr 10;14(4):evac045. doi: 10.1093/gbe/evac045. Genome Biol Evol. 2022. PMID: 35445261 Free PMC article. - Inferences of African evolutionary history from genomic data.
Beltrame MH, Rubel MA, Tishkoff SA. Beltrame MH, et al. Curr Opin Genet Dev. 2016 Dec;41:159-166. doi: 10.1016/j.gde.2016.10.002. Epub 2016 Nov 1. Curr Opin Genet Dev. 2016. PMID: 27810637 Free PMC article. Review. - Approximate Bayesian computation with deep learning supports a third archaic introgression in Asia and Oceania.
Mondal M, Bertranpetit J, Lao O. Mondal M, et al. Nat Commun. 2019 Jan 16;10(1):246. doi: 10.1038/s41467-018-08089-7. Nat Commun. 2019. PMID: 30651539 Free PMC article.
References
- Bandelt HJ, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16: 37–48. - PubMed
- Bräuer G. 2008. The origin of modern anatomy: by speciation or intraspecific evolution? Evol Anthropol 17: 22–37.
- Campana MG, Bower MA, Crabtree PJ. 2013. Ancient DNA for the archaeologist: the future of African research. Afr Archaeol Rev 30: 21–37.
- Cavalli-Sforza LL, Menozzi P, Piazza A. 1994. The history and geography of human genes. Princeton University Press, Princeton, NJ.
Publication types
MeSH terms
Grants and funding
- 8DP1ES022577-04/DP/NCCDPHP CDC HHS/United States
- R01 HG005226/HG/NHGRI NIH HHS/United States
- 1R01GM113657-01/GM/NIGMS NIH HHS/United States
- F32HG006648/HG/NHGRI NIH HHS/United States
- R01 GM113657/GM/NIGMS NIH HHS/United States
- F32 HG006648/HG/NHGRI NIH HHS/United States
- DP1 ES022577/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources