PARP1 Links CHD2-Mediated Chromatin Expansion and H3.3 Deposition to DNA Repair by Non-homologous End-Joining - PubMed (original) (raw)
PARP1 Links CHD2-Mediated Chromatin Expansion and H3.3 Deposition to DNA Repair by Non-homologous End-Joining
Martijn S Luijsterburg et al. Mol Cell. 2016.
Abstract
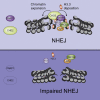
The response to DNA double-strand breaks (DSBs) requires alterations in chromatin structure to promote the assembly of repair complexes on broken chromosomes. Non-homologous end-joining (NHEJ) is the dominant DSB repair pathway in human cells, but our understanding of how it operates in chromatin is limited. Here, we define a mechanism that plays a crucial role in regulating NHEJ in chromatin. This mechanism is initiated by DNA damage-associated poly(ADP-ribose) polymerase 1 (PARP1), which recruits the chromatin remodeler CHD2 through a poly(ADP-ribose)-binding domain. CHD2 in turn triggers rapid chromatin expansion and the deposition of histone variant H3.3 at sites of DNA damage. Importantly, we find that PARP1, CHD2, and H3.3 regulate the assembly of NHEJ complexes at broken chromosomes to promote efficient DNA repair. Together, these findings reveal a PARP1-dependent process that couples ATP-dependent chromatin remodeling with histone variant deposition at DSBs to facilitate NHEJ and safeguard genomic stability.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
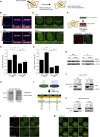
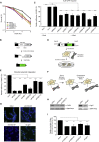
Chromatin Changes in Response to DNA Damage Depend on PARP1 (A) Outline of the chromatin expansion approach. (B) PAGFP-H2A expansion in U2OS cells treated with DMSO or 10 μM PARPi. (C) Quantification of (B). (D) GFP-XRCC4 expansion in U2OS cells treated with DMSO or 10 μM PARPi. (E) Quantification of (D). 40–65 cells were analyzed from three independent experiments. (F) Outline of the chromatin-tethering approach in U2OS 2-6-3 cells, which identified CHD2 as a PARP1 interactor. (G) CoIP of endogenous PARP1 and CHD2 in HEK293 cells. (H) Western blot on U2OS CHD2-GFP cells. (I) SILAC of HEK293 cells expressing GFP (L) or CHD2-GFP (H). (J) CoIP of CHD2-GFP and endogenous PARP1 in HEK293 cells. (K) PARylation of CHD2-GFP in HEK293 cells. (L) Recruitment of endogenous CHD2 to UV-A tracks in U2OS cells. CHD2 knockdown confirms antibody specificity. (M) Recruitment of CHD2-GFP to multi-photon tracks in U2OS cells.
Figure 2
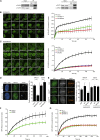
CHD2 Recruitment to DNA Damage Requires PARP1 (A) Recruitment of CHD2-GFP to multi-photon tracks in U2OS cells treated with DMSO or 1 μM PARPi. (B) Quantification of (A). (C) Recruitment of endogenous CHD2 in U2OS cells treated with DMSO or 10 μM PARPi. (D) Recruitment of CHD2-GFP in cells transfected with the indicated siRNAs. (E) Western blot showing PARP1/2 knockdown efficiency in cells from (D). (F) Quantification of (D). 30–170 cells were analyzed from three independent experiments.
Figure 3
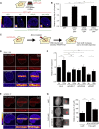
The C Terminus of CHD2 Is Required for DNA Damage Recruitment and PAR Binding (A) Schematic representation of CHD2 and its deletion mutants. (B) Western blot showing expression of the mutants from A in U2OS cells. (C) Recruitment of CHD2-GFP deletion mutants in cells depleted for endogenous CHD2 by siCHD2-69 or siCHD2-17 in case of CHD2952–1391-GFP. CHD21–1611-GFP was rendered siCHD2-69-resistant. NBS1-mCherry was a DNA damage marker. (D) Quantification of (C). 10–30 cells were analyzed from two independent experiments. (E) IP of the indicated CHD2-GFP fragments from HEK293 cells followed by southwestern blotting to monitor association with radiolabeled PAR. Recombinant PARP1 was a control. CoIP with endogenous PARP1 is shown in the bottom panel. (F) Quantification of (E) and two additional independent experiments. PAR binding levels of CHD2WT-GFP were set to 1.
Figure 4
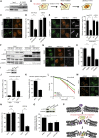
CHD2 Promotes DSB Repair by NHEJ (A) Clonogenic survival after IR exposure of VH10-SV40 cells transfected with the indicated siRNAs. (B) Schematic representation of the EJ5-GFP reporter for NHEJ. (C) Quantification of EJ5-GFP-positive HEK293 cells corrected for I-SceI transfection efficiency by co-transfection with mCherry. The average of four independent experiments is shown. (D) Schematic representation of the plasmid integration assay. (E) Quantification of the plasmid integration efficiency in U2OS cells from three independent experiments. (F) TRF2ts MEFs were shifted to the non-permissive temperature to induce telomere uncapping and NHEJ-dependent chromosome fusions. (G) Representative images of metaphases from TRF2ts MEFs transduced with the indicated shRNAs after 24 hr of telomere uncapping. Telomere-FISH shows the position of the telomeres (green), and chromosomes are stained by DAPI (blue). (H) Western blot showing CHD2 and LigIV knockdown efficiency in TRF2ts MEFs. (I) Quantification of interchromosomal fusions observed in cells transduced with the indicated shRNAs. Scrambled control shRNA (shScr) was normalized to 100%. 4500–8000 chromosomes were analyzed from three to eight independent experiments.
Figure 5
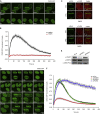
CHD2 Promotes the Assembly of NHEJ Complexes at DSBs (A) Reciprocal coIPs of endogenous CHD2 and KU70 in HEK293 cells. (B) Recruitment of GFP-KU70 to multi-photon tracks in HeLa cells. (C) As in (B), except for GFP-XRCC4 in U2OS cells. (D) siRNA-transfected pTuner265 cells were induced for FokI-LacR expression and stained for γH2AX and XRCC4. (E) U2OS cells transfected with the indicated constructs were UV-A micro-irradiated and stained for γH2AX and XRCC4. (F and G) Recruitment kinetics of GFP-XRCC4 to multi-photon tracks in (F) U2OS cells treated with DMSO or 10 μM PARPi or in (G) U2OS cells transfected with the indicated siRNAs. 40–160 cells were analyzed from (B) three or (C–E) two independent experiments.
Figure 6
CHD2 Promotes DNA Damage-Induced Chromatin Changes (A) U2OS 2-6-3 cells containing a LacO array were co-transfected with αGFP-mCherry-LacR and GFP-tagged CHD2 variants or GFP only. (B) Quantification of the array size upon tethering of the indicated proteins. (C) Outline of the PAGFP-H2A expansion approach. (D and E) PAGFP-H2A expansion in U2OS cells transfected with the indicated siRNAs. NBS1-mCherry was a DNA damage marker. (F) Quantification of the PAGFP-H2A expansion experiments. Values were corrected for the expansion in fixed cells and normalized to 1 for siLuc at 5 s post irradiation. (G) U2OS 2-6-3 cells were transfected with mCherry-LacR, whereas 2-6-3-derived pTuner265 cells were transfected with the indicated siRNAs and induced for FokI-LacR expression. 60–100 cells were analyzed from (B and F) three or (G) two independent experiments.
Figure 7
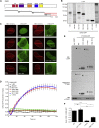
CHD2-Mediated H3.3 Deposition Regulates NHEJ (A) CoIP of endogenous CHD2 and H3.3 in HEK293 cells. (B) Outline of the H3.3 deposition approach. (C) Deposition of H3.3 at UV-A tracks in U2OS cells transfected with the indicated siRNAs. (D) Quantification of (C). (E) U2OS H3.3-SNAP cells transfected with the indicated GFP constructs were UV-A micro-irradiated, SNAP-labeled, and stained for γH2AX. (F) CoIP of GFP-H3.3 and KU80 or PARP1 in U2OS cells. (G) Deposition of H3.3 at UV-A tracks in U2OS cells treated with DMSO or 10 μM PARPi. (H) Quantification of (G) and of cells transfected with PARP1 siRNAs. (I) Western blot showing H3.3 knockdown efficiency in U2OS cells. (J) Quantification of GFP-positive EJ5-GFP HEK293 cells corrected for I-SceI transfection efficiency by co-transfection with mCherry. The average of two independent experiments is shown. (K) Plasmid integration assay after H3.3 knockdown in U2OS cells. The average of two independent experiments is shown. (L) Clonogenic survival after IR exposure of siRNA-transfected VH10-SV40 cells. (M) XRCC4 recruitment to UV-A tracks in siRNA-transfected U2OS cells. (N) Quantification of (M). 45–100 cells were analyzed from (E) two or (D and H) three independent experiments. (O) Quantification of interchromosomal fusions observed in shRNA-transduced TRF2ts MEFs after telomere uncapping. 3,200–4,600 chromosomes were analyzed from two independent experiments. Western blot showing H3.3 knockdown efficiency. (P) Model for how PARP1 links CHD2-mediated chromatin expansion and H3.3 assembly to DSB repair by NHEJ.
Similar articles
- The absence of Ku but not defects in classical non-homologous end-joining is required to trigger PARP1-dependent end-joining.
Mansour WY, Borgmann K, Petersen C, Dikomey E, Dahm-Daphi J. Mansour WY, et al. DNA Repair (Amst). 2013 Dec;12(12):1134-42. doi: 10.1016/j.dnarep.2013.10.005. Epub 2013 Nov 7. DNA Repair (Amst). 2013. PMID: 24210699 - Common and unique genetic interactions of the poly(ADP-ribose) polymerases PARP1 and PARP2 with DNA double-strand break repair pathways.
Ghosh R, Roy S, Kamyab J, Danzter F, Franco S. Ghosh R, et al. DNA Repair (Amst). 2016 Sep;45:56-62. doi: 10.1016/j.dnarep.2016.06.001. Epub 2016 Jun 16. DNA Repair (Amst). 2016. PMID: 27373144 Free PMC article. - Histone deacetylase inhibitors decrease NHEJ both by acetylation of repair factors and trapping of PARP1 at DNA double-strand breaks in chromatin.
Robert C, Nagaria PK, Pawar N, Adewuyi A, Gojo I, Meyers DJ, Cole PA, Rassool FV. Robert C, et al. Leuk Res. 2016 Jun;45:14-23. doi: 10.1016/j.leukres.2016.03.007. Epub 2016 Mar 30. Leuk Res. 2016. PMID: 27064363 Free PMC article. - Touching base with PARPs: moonlighting in the repair of UV lesions and double-strand breaks.
Pines A, Mullenders LH, van Attikum H, Luijsterburg MS. Pines A, et al. Trends Biochem Sci. 2013 Jun;38(6):321-30. doi: 10.1016/j.tibs.2013.03.002. Epub 2013 Apr 4. Trends Biochem Sci. 2013. PMID: 23562323 Review. - Relation between carcinogenesis, chromatin structure and poly(ADP-ribosylation) (review).
Boulikas T. Boulikas T. Anticancer Res. 1991 Mar-Apr;11(2):489-527. Anticancer Res. 1991. PMID: 1905900 Review.
Cited by
- Real-Time Tracking of Parental Histones Reveals Their Contribution to Chromatin Integrity Following DNA Damage.
Adam S, Dabin J, Chevallier O, Leroy O, Baldeyron C, Corpet A, Lomonte P, Renaud O, Almouzni G, Polo SE. Adam S, et al. Mol Cell. 2016 Oct 6;64(1):65-78. doi: 10.1016/j.molcel.2016.08.019. Epub 2016 Sep 15. Mol Cell. 2016. PMID: 27642047 Free PMC article. - Hypermethylation of RAD9A intron 2 in childhood cancer patients, leukemia and tumor cell lines suggest a role for oncogenic transformation.
Galetzka D, Böck J, Wagner L, Dittrich M, Sinizyn O, Ludwig M, Rossmann H, Spix C, Radsak M, Scholz-Kreisel P, Mirsch J, Linke M, Brenner W, Marron M, Poplawski A, Haaf T, Schmidberger H, Prawitt D. Galetzka D, et al. EXCLI J. 2022 Jan 7;21:117-143. doi: 10.17179/excli2021-4482. eCollection 2022. EXCLI J. 2022. PMID: 35221838 Free PMC article. - The comings and goings of PARP-1 in response to DNA damage.
Pascal JM. Pascal JM. DNA Repair (Amst). 2018 Nov;71:177-182. doi: 10.1016/j.dnarep.2018.08.022. Epub 2018 Aug 23. DNA Repair (Amst). 2018. PMID: 30177435 Free PMC article. Review. - Chk1 Inhibition Hinders the Restoration of H3.1K56 and H3.3K56 Acetylation and Reprograms Gene Transcription After DNA Damage Repair.
Ding N, Shao Z, Yuan F, Qu P, Li P, Lu D, Wang J, Zhu Q. Ding N, et al. Front Oncol. 2022 Apr 14;12:862592. doi: 10.3389/fonc.2022.862592. eCollection 2022. Front Oncol. 2022. PMID: 35494003 Free PMC article. - Pleiotropic role of PARP1: an overview.
Kumar V, Kumar A, Mir KUI, Yadav V, Chauhan SS. Kumar V, et al. 3 Biotech. 2022 Jan;12(1):3. doi: 10.1007/s13205-021-03038-6. Epub 2021 Dec 4. 3 Biotech. 2022. PMID: 34926116 Free PMC article. Review.
References
- Adam S., Polo S.E., Almouzni G. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell. 2013;155:94–106. - PubMed
- Audebert M., Salles B., Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279:55117–55126. - PubMed
- Boehler C., Gauthier L.R., Mortusewicz O., Biard D.S., Saliou J.M., Bresson A., Sanglier-Cianferani S., Smith S., Schreiber V., Boussin F., Dantzer F. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc. Natl. Acad. Sci. USA. 2011;108:2783–2788. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous