The Plant Heat Stress Transcription Factors (HSFs): Structure, Regulation, and Function in Response to Abiotic Stresses - PubMed (original) (raw)
Review
The Plant Heat Stress Transcription Factors (HSFs): Structure, Regulation, and Function in Response to Abiotic Stresses
Meng Guo et al. Front Plant Sci. 2016.
Abstract
Abiotic stresses such as high temperature, salinity, and drought adversely affect the survival, growth, and reproduction of plants. Plants respond to such unfavorable changes through developmental, physiological, and biochemical ways, and these responses require expression of stress-responsive genes, which are regulated by a network of transcription factors (TFs), including heat stress transcription factors (HSFs). HSFs play a crucial role in plants response to several abiotic stresses by regulating the expression of stress-responsive genes, such as heat shock proteins (Hsps). In this review, we describe the conserved structure of plant HSFs, the identification of HSF gene families from various plant species, their expression profiling under abiotic stress conditions, regulation at different levels and function in abiotic stresses. Despite plant HSFs share highly conserved structure, their remarkable diversification across plants reflects their numerous functions as well as their integration into the complex stress signaling and response networks, which can be employed in crop improvement strategies via biotechnological intervention.
Keywords: abiotic stress; heat shock proteins; heat stress; plant; transcription factors; transcriptional regulation.
Figures
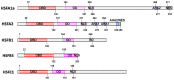
Figure 1
Basic structure of HSFs. The block diagrams represent five tomato HSFs with their conserved functional domains. The conserved domains are identified by Heatster (
http://www.cibiv.at/services/hsf/
). DBD, DNA binding domain; OD, oligomerization domain (HR-A/B region); NLS, nuclear localization signal; NES, nuclear export signal; AHA, activator motifs; RD, tetrapeptide motif–LFGV–as core of repressor domain. (Adapted from Scharf et al., 2012).
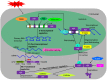
Figure 2
Regulation of HSF proteins. The scheme depicts the regulation of HSFs at different levels during stress. Upstream TFs like DREB, HSF, or ABI may bind to stress-related _cis_-regulatory elements in the promoter of regulated HSF genes and influence their transcription. Post-transcriptional control of HSFs by alternative splicing may also regulate their expression. The mature mRNAs are again governed during their transport and translation. uORFs regulate HSFs at the translation level. The translated protein may be subjected to activation by phosphorylation or undergo SUMO- and ubiquitin proteasomal-mediated degradation in response to certain environmental cues, other translated HSF proteins may be sequestrated by their inhibitors. Upon their nuclear import, the activated HSF proteins homo- or heterodimerize or bind to promoters of their target genes to control their expression. Broken arrows indicate possible but not firmly demonstrated routes. The red X mark represents translational repression. DREB, dehydration responsive element binding protein; ABI, ABSCISIC ACID–INSENSITIVE protein; TFs, transcription factors; AS, alternate splicing; mRNA, messenger RNA; m7G, cap of mRNA; uORFs, upstream micro open reading frames; mORF, major ORF; uAUG, AUG of uORF; mAUG, AUG of mORF; P, phosphate; SUMO, small ubiquitin-like modifier; Ubi, ubiquitination; HSE, heat stress element. (Adapted from Calkhoven and Ab, ; Puranik et al., 2012).
Figure 3
Schematic representation of HSFs as key components in transcriptional regulatory networks during abiotic stress. The scheme integrates both positive (arrows) and negative (bars) regulatory mechanisms. Abiotic stresses provoke a rise of cytoplasmic calcium, ROS accumulation and proteins denaturation inside the cells which convey stress-induced signals to responding genes, directly targeting HSF proteins marked with an asterisk. HSFs induce the activation of various genes playing a central role under abiotic stress conditions, thereby enhancing the abiotic stress tolerance. ROS, reactive oxygen species; CaM, Ca2+–calmodulin; TFs, transcription factors; Hsp, heat shock protein; sHsp, small Hsp; HSE, heat stress element; Hsa32, heat stress- associated 32-kD protein; Rof1, FK506-binding proteins; GST, glutathione-S-transferase; RD29A, drought-regulated gene 29A; APX2, ascorbate peroxidase 2; GolS1, a galactinol synthase; HSBP, HSF binding protein.
Similar articles
- Genome-wide identification and transcript profiles of walnut heat stress transcription factor involved in abiotic stress.
Liu X, Meng P, Yang G, Zhang M, Peng S, Zhai MZ. Liu X, et al. BMC Genomics. 2020 Jul 10;21(1):474. doi: 10.1186/s12864-020-06879-2. BMC Genomics. 2020. PMID: 32650719 Free PMC article. - Genome-wide characterization and evolutionary analysis of heat shock transcription factors (HSFs) to reveal their potential role under abiotic stresses in radish (Raphanus sativus L.).
Tang M, Xu L, Wang Y, Cheng W, Luo X, Xie Y, Fan L, Liu L. Tang M, et al. BMC Genomics. 2019 Oct 24;20(1):772. doi: 10.1186/s12864-019-6121-3. BMC Genomics. 2019. PMID: 31651257 Free PMC article. - [Biological characteristics of heat shock transcription factors and their roles in abiotic stress adaptation of higher plant].
Shao KZ, Lyu XP, Li JL, Chen J, Zhao LY, Ren W, Zhang JL. Shao KZ, et al. Ying Yong Sheng Tai Xue Bao. 2022 Aug;33(8):2286-2296. doi: 10.13287/i.1001-9332.202208.039. Ying Yong Sheng Tai Xue Bao. 2022. PMID: 36043838 Review. Chinese. - [Heat Shock Proteins in Plant Protection from Oxidative Stress].
Yurina NP. Yurina NP. Mol Biol (Mosk). 2023 Nov-Dec;57(6):949-964. Mol Biol (Mosk). 2023. PMID: 38062952 Review. Russian.
Cited by
- Plant responses to high temperature and drought: A bibliometrics analysis.
Cui Y, Ouyang S, Zhao Y, Tie L, Shao C, Duan H. Cui Y, et al. Front Plant Sci. 2022 Nov 9;13:1052660. doi: 10.3389/fpls.2022.1052660. eCollection 2022. Front Plant Sci. 2022. PMID: 36438139 Free PMC article. - Tomato plant response to heat stress: a focus on candidate genes for yield-related traits.
Graci S, Barone A. Graci S, et al. Front Plant Sci. 2024 Jan 8;14:1245661. doi: 10.3389/fpls.2023.1245661. eCollection 2023. Front Plant Sci. 2024. PMID: 38259925 Free PMC article. Review. - Untranslated yet indispensable-UTRs act as key regulators in the environmental control of gene expression.
Hardy EC, Balcerowicz M. Hardy EC, et al. J Exp Bot. 2024 Jul 23;75(14):4314-4331. doi: 10.1093/jxb/erae073. J Exp Bot. 2024. PMID: 38394144 Free PMC article. Review. - Genome-wide identification and transcript profiles of walnut heat stress transcription factor involved in abiotic stress.
Liu X, Meng P, Yang G, Zhang M, Peng S, Zhai MZ. Liu X, et al. BMC Genomics. 2020 Jul 10;21(1):474. doi: 10.1186/s12864-020-06879-2. BMC Genomics. 2020. PMID: 32650719 Free PMC article. - Transcriptional regulation of osmotic stress tolerance in wheat (Triticum aestivum L.).
Wani SH, Tripathi P, Zaid A, Challa GS, Kumar A, Kumar V, Upadhyay J, Joshi R, Bhatt M. Wani SH, et al. Plant Mol Biol. 2018 Aug;97(6):469-487. doi: 10.1007/s11103-018-0761-6. Epub 2018 Aug 14. Plant Mol Biol. 2018. PMID: 30109563 Review.
References
- Al-Whaibi M. H. (2011). Plant heat-shock proteins: a mini review. J. King Saud Univ.-Sci. 23, 139–150. 10.1016/j.jksus.2010.06.022 - DOI
- Bartels D., Sunkar R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24, 23–58. 10.1080/07352680590910410 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources