Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children - PubMed (original) (raw)
. 2016 Feb 19;351(6275):10.1126/science.aad3311 aad3311.
doi: 10.1126/science.aad3311.
Mark R Charbonneau 1, Tarek Salih 1, Michael J Barratt 1, Siddarth Venkatesh 1, Olga Ilkaveya 2, Sathish Subramanian 1, Mark J Manary 3, Indi Trehan 4, Josh M Jorgensen 5, Yue-Mei Fan 6, Bernard Henrissat 7, Semen A Leyn 8, Dmitry A Rodionov 9, Andrei L Osterman 10, Kenneth M Maleta 11, Christopher B Newgard 12, Per Ashorn 13, Kathryn G Dewey 5, Jeffrey I Gordon 1
Affiliations
- PMID: 26912898
- PMCID: PMC4787260
- DOI: 10.1126/science.aad3311
Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children
Laura V Blanton et al. Science. 2016.
Abstract
Undernourished children exhibit impaired development of their gut microbiota. Transplanting microbiota from 6- and 18-month-old healthy or undernourished Malawian donors into young germ-free mice that were fed a Malawian diet revealed that immature microbiota from undernourished infants and children transmit impaired growth phenotypes. The representation of several age-discriminatory taxa in recipient animals correlated with lean body mass gain; liver, muscle, and brain metabolism; and bone morphology. Mice were cohoused shortly after receiving microbiota from healthy or severely stunted and underweight infants; age- and growth-discriminatory taxa from the microbiota of the former were able to invade that of the latter, which prevented growth impairments in recipient animals. Adding two invasive species, Ruminococcus gnavus and Clostridium symbiosum, to the microbiota from undernourished donors also ameliorated growth and metabolic abnormalities in recipient animals. These results provide evidence that microbiota immaturity is causally related to undernutrition and reveal potential therapeutic targets and agents.
Copyright © 2016, American Association for the Advancement of Science.
Figures
Fig. 1. Sparse Random Forests-derived model of gut microbiota maturation obtained from concordant healthy Malawian twins/triplets
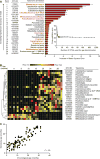
(A) Random-Forests regression of fecal bacterial 97%ID OTUs from a training set of healthy Malawian infants/children (_n_=31) to chronological age yielded a rank order of age-discriminatory taxa. Random Forests assigns a mean squared error (MSE), or feature importance score, to each OTU that indicates the extent to which each OTU contributes to the accuracy of the model. The 25 most age-discriminatory taxa, ranked by MSE, yielded a sparse model that predicted microbiota age and accounted for ~80% of the observed variance in the healthy cohort (see Table S3A for a complete list of OTUs and MSE values). The top 25 most discriminatory OTUs with their taxonomic assignments are shown ranked by feature importance (mean±SD of the MSE). The insert shows the results of 10-fold cross-validation; as OTUs are added to the model in order of their feature importance rank, the model’s error decreases. Taxa highlighted in red indicate OTUs with >97% nucleotide sequence identity with an OTU present in a sparse Random Forests-based model of microbiota maturation in healthy Bangladeshi infants/children (11). (B) A heatmap of changes over time in relative abundances of the 25 OTUs in fecal microbiota collected from healthy Malawian infants/children comprising the test set (_n_=29). OTUs are hierarchically clustered according to pairwise distances by Pearson correlation. (C) Predictions of chronological age using the sparse 25 OTU model of microbiota age in healthy children comprising the test set cohort. r2 calculated from Pearson correlation.
Fig. 2. Transplantation of microbiota from 6- and 18-month old donors to young germ-free mice provides evidence of a causal relationship between gut microbiota maturity and growth phenotypes
(A) Experimental design of microbiota screen. Mice (4.5-weeks old) were switched to the M8 diet three days prior to gavage with the selected microbiota donor’s fecal sample (_n_=5 mice per donor). Fecal samples, body weight and body composition were defined at the indicated time. (B,C) Gnotobiotic mice colonized with fecal samples from healthy children gain more total body weight (panel B) and lean mass (panel C) than mice colonized with microbiota from undernourished donors (mean±SEM shown; p-values shown for donor status effect based on 2-way ANOVA). All recipient mice harbor microbiota that represent >50% of OTU diversity present in the intact uncultured donor’s sample. (D) The 30 most weight-gain discriminatory OTUs and their taxonomic assignments, ranked by feature importance (mean MSE±SD values are plotted). The weight gain model explained ~66% of the observed phenotypic variation (p<0.0001, permutation test; 999 permutations). Taxa in red indicate OTUs that appear within the 30 most discriminatory OTUs for both the weight and lean mass gain Random Forests-based models. Taxa in purple indicate species that appear in the 25-member sparse Random Forests-derived model of Malawian gut microbiota maturation. Bars to the right of the OTU ID numbers represent Spearman’s rank correlation of the same OTU ID to chronological age within the healthy Malawian infant/child cohort (see Table S9).
Fig. 3. Co-housing results in transfer of species from the microbiota of cagemates colonized with the healthy donor’s community into the microbiota of cagemates containing the severely stunted/underweight donor’s community and prevention of growth faltering
(A) Experimental design for co-housing experiments. Dually housed 4.5-week old mice were switched to the M8 diet and colonized 3 days later with either the intact uncultured healthy or stunted/underweight donor microbiota. Four days after gavage, subsets of the mice were co-housed (HCH and UnCH, respectively), while healthy and stunted/underweight control mice remained in their original isolators and were paired with a new cagemate from that isolator (H-H and Un-Un controls). Fecal samples were collected throughout the experiment; growth was assayed by changes in total body weight and body composition (the latter by qMR). Mice were sacrificed three weeks after colonization. (B) HCH and UnCH mice have increased lean mass gain relative to the Un-Un controls 15 days post colonization (Un-Un vs. HCH p = 0.0447, Un-Un vs. UnCH p=0.0121, Mann-Whitney test; _n_=6 cages of co-housed mice, 3 cages of each dually housed control group/experiment; two independent experiments). To quantify invasion further, we used the mean and standard deviation of the null distribution of invasion scores (defined as the scores from recipients of the H or Un microbiota that had never been co-housed with each other) to calculate a z-value and a Benjamini-Hochberg adjusted p-value for the invasion score of each species in HCH and UnCH mice (see Materials and Methods). We defined a taxon as a successful invader if it (i) had a Benjamini-Hochberg adjusted p≤0.05, and (ii) had a relative abundance of ≤0.05% before cohousing and ≥0.5% in the fecal microbiota at the time of sacrifice. Fig. 3C and Table S12 provide information about the direction and success of invasion. (C) Heatmap showing results of the invasion assay. Each row represents a species-level taxon, while each column represents a mouse at a given day post colonization (dpc); rows of the heatmap were hierarchically clustered according to pair-wise distances using Pearson correlation. Bars at the right side of each experimental arm represent the fold-change (fc) in that species’ relative abundance before and after cohousing (fold-change defined by the log2[(average relative abundance of species post cohousing (days 7 through 22))/(average relative abundance of species before cohousing (day 4))]. Species in red represent those identified as one of the top 30 growth-discriminatory taxa by the weight or lean mass gain Random Forests-based models shown in Fig. 2.
Fig. 4. A consortium of cultured growth-discriminatory OTUs augments the growth of mice colonized with the 6-month severely stunted/underweight donor’s microbiota
(A) Experimental design including the composition of the 5-member consortium of cultured bacterial strains. (B) Weight gain and lean body mass gain 21 days after gavage of the donor microbiota with or without the cultured consortium. (C) Comparison of the fecal microbiota of mice belonging to untreated control and treated experimental group showing the establishment of OTUs from the consortium at 21 days post colonization. (D) The effects of treatment with the consortium on host metabolism (p<0.05 for all metabolites shown, Student’s t-test). Each row represents a metabolite from a given tissue, while each column represents an individual mouse. Tissues were collected 21 days after colonization.
Comment in
- Microbiome: Restoring healthy growth in infants.
Du Toit A. Du Toit A. Nat Rev Microbiol. 2016 Apr;14(4):191. doi: 10.1038/nrmicro.2016.31. Epub 2016 Feb 29. Nat Rev Microbiol. 2016. PMID: 26923113 No abstract available. - Gut microbiota: Growth impairment in undernourished children.
Greenhill C. Greenhill C. Nat Rev Endocrinol. 2016 Apr;12(4):186. doi: 10.1038/nrendo.2016.34. Epub 2016 Mar 4. Nat Rev Endocrinol. 2016. PMID: 26939979 No abstract available. - Gut microbiota: How to build healthy growth-promoting gut communities.
Relman DA. Relman DA. Nat Rev Gastroenterol Hepatol. 2016 Jul;13(7):379-80. doi: 10.1038/nrgastro.2016.74. Epub 2016 May 18. Nat Rev Gastroenterol Hepatol. 2016. PMID: 27188819 No abstract available. - Healthy gut microbiota can resolve undernutrition.
Hermann E, Foligne B. Hermann E, et al. Hepatobiliary Surg Nutr. 2017 Apr;6(2):141-143. doi: 10.21037/hbsn.2017.01.19. Hepatobiliary Surg Nutr. 2017. PMID: 28503565 Free PMC article. No abstract available.
Similar articles
- Sialylated Milk Oligosaccharides Promote Microbiota-Dependent Growth in Models of Infant Undernutrition.
Charbonneau MR, O'Donnell D, Blanton LV, Totten SM, Davis JC, Barratt MJ, Cheng J, Guruge J, Talcott M, Bain JR, Muehlbauer MJ, Ilkayeva O, Wu C, Struckmeyer T, Barile D, Mangani C, Jorgensen J, Fan YM, Maleta K, Dewey KG, Ashorn P, Newgard CB, Lebrilla C, Mills DA, Gordon JI. Charbonneau MR, et al. Cell. 2016 Feb 25;164(5):859-71. doi: 10.1016/j.cell.2016.01.024. Epub 2016 Feb 18. Cell. 2016. PMID: 26898329 Free PMC article. - Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy.
Kau AL, Planer JD, Liu J, Rao S, Yatsunenko T, Trehan I, Manary MJ, Liu TC, Stappenbeck TS, Maleta KM, Ashorn P, Dewey KG, Houpt ER, Hsieh CS, Gordon JI. Kau AL, et al. Sci Transl Med. 2015 Feb 25;7(276):276ra24. doi: 10.1126/scitranslmed.aaa4877. Sci Transl Med. 2015. PMID: 25717097 Free PMC article. - Gut microbiota: Growth impairment in undernourished children.
Greenhill C. Greenhill C. Nat Rev Endocrinol. 2016 Apr;12(4):186. doi: 10.1038/nrendo.2016.34. Epub 2016 Mar 4. Nat Rev Endocrinol. 2016. PMID: 26939979 No abstract available. - Gut Microbiome and Bone: to Build, Destroy, or Both?
Yan J, Charles JF. Yan J, et al. Curr Osteoporos Rep. 2017 Aug;15(4):376-384. doi: 10.1007/s11914-017-0382-z. Curr Osteoporos Rep. 2017. PMID: 28620867 Free PMC article. Review. - Feeding the brain and nurturing the mind: Linking nutrition and the gut microbiota to brain development.
Goyal MS, Venkatesh S, Milbrandt J, Gordon JI, Raichle ME. Goyal MS, et al. Proc Natl Acad Sci U S A. 2015 Nov 17;112(46):14105-12. doi: 10.1073/pnas.1511465112. Proc Natl Acad Sci U S A. 2015. PMID: 26578751 Free PMC article. Review.
Cited by
- Dairy and plant based food intakes are associated with altered faecal microbiota in 2 to 3 year old Australian children.
Smith-Brown P, Morrison M, Krause L, Davies PS. Smith-Brown P, et al. Sci Rep. 2016 Oct 3;6:32385. doi: 10.1038/srep32385. Sci Rep. 2016. PMID: 27694811 Free PMC article. - A metagenome-wide association study of gut microbiome and visceral fat accumulation.
Nie X, Chen J, Ma X, Ni Y, Shen Y, Yu H, Panagiotou G, Bao Y. Nie X, et al. Comput Struct Biotechnol J. 2020 Sep 20;18:2596-2609. doi: 10.1016/j.csbj.2020.09.026. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 33033580 Free PMC article. - Gut Microbiota, Muscle Mass and Function in Aging: A Focus on Physical Frailty and Sarcopenia.
Ticinesi A, Nouvenne A, Cerundolo N, Catania P, Prati B, Tana C, Meschi T. Ticinesi A, et al. Nutrients. 2019 Jul 17;11(7):1633. doi: 10.3390/nu11071633. Nutrients. 2019. PMID: 31319564 Free PMC article. Review. - Microbial Control of Intestinal Homeostasis via Enteroendocrine Cell Innate Immune Signaling.
Watnick PI, Jugder BE. Watnick PI, et al. Trends Microbiol. 2020 Feb;28(2):141-149. doi: 10.1016/j.tim.2019.09.005. Epub 2019 Nov 4. Trends Microbiol. 2020. PMID: 31699645 Free PMC article. Review. - Effects of feeding Saccharomyces cerevisiae fermentation postbiotic on the fecal microbial community of Holstein dairy calves.
Centeno-Martinez RE, Dong W, Klopp RN, Yoon I, Boerman JP, Johnson TA. Centeno-Martinez RE, et al. Anim Microbiome. 2023 Feb 19;5(1):13. doi: 10.1186/s42523-023-00234-y. Anim Microbiome. 2023. PMID: 36803311 Free PMC article.
References
- Black RE, et al. Maternal and child undernutrition and overweight in low-income and middle-income countries. Lancet. 2013;382:427–451. - PubMed
MeSH terms
Grants and funding
- R01 DK030292/DK/NIDDK NIH HHS/United States
- T32 AI007172/AI/NIAID NIH HHS/United States
- P30 AR057235/AR/NIAMS NIH HHS/United States
- T32 GM007067/GM/NIGMS NIH HHS/United States
- R37 DK030292/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical