Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences - PubMed (original) (raw)
Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences
Charlotte Soneson et al. F1000Res. 2015.
Abstract
High-throughput sequencing of cDNA (RNA-seq) is used extensively to characterize the transcriptome of cells. Many transcriptomic studies aim at comparing either abundance levels or the transcriptome composition between given conditions, and as a first step, the sequencing reads must be used as the basis for abundance quantification of transcriptomic features of interest, such as genes or transcripts. Various quantification approaches have been proposed, ranging from simple counting of reads that overlap given genomic regions to more complex estimation of underlying transcript abundances. In this paper, we show that gene-level abundance estimates and statistical inference offer advantages over transcript-level analyses, in terms of performance and interpretability. We also illustrate that the presence of differential isoform usage can lead to inflated false discovery rates in differential gene expression analyses on simple count matrices but that this can be addressed by incorporating offsets derived from transcript-level abundance estimates. We also show that the problem is relatively minor in several real data sets. Finally, we provide an R package ( tximport) to help users integrate transcript-level abundance estimates from common quantification pipelines into count-based statistical inference engines.
Keywords: RNA-seq; gene expression; quantification; transcriptomics.
Conflict of interest statement
Competing interests: No competing interests were disclosed.
Figures
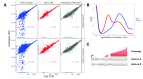
Figure 1 (sim2).
A: Accuracy of gene- and transcript-level TPM estimates from_Salmon_ and scaled FPKM estimates derived from simple counts from_featureCounts_, in one of the simulated samples (sampleA1). Spearman correlations are indicated in the respective panels. Top row: using the complete annotation. Bottom row: using an incomplete annotation, with 20% of the transcripts randomly removed. Gene-level estimates are more accurate than transcript-level estimates. Gene-level estimates from_Salmon_ are more accurate than those from_featureCounts_.B: Distribution of the coefficients of variation of gene- and transcript-level abundance estimates from_Salmon_, calculated across 30 bootstrap samples of one of the simulated samples (sampleA1). Gene-level estimates are less variable than transcript-level estimates.C: An example of unidentifiable transcript-level estimates, as uneven coverage does not cover the critical regions that would determine the amount that each transcript is expressed, while gene-level estimation is still possible.
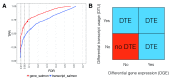
Figure 2 (sim2).
A: DTE detection performance on transcript- and gene-level, using_edgeR_ applied to transcript-level estimated counts from_Salmon_. The statistical analysis was performed on transcript level and aggregated for each gene using the_perGeneQValue_ function from the_DEXSeq_ R package; aggregated results show higher detection power. The curves trace out the observed FDR and TPR for each significance cutoff value. The three circles mark the performance at adjusted p-value cutoffs of 0.01, 0.05 and 0.1.B: Schematic illustration of different ways in which differential transcript expression (DTE) can arise, in terms of absence or presence of differential gene expression (DGE) and differential transcript usage (DTU).
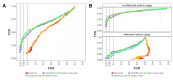
Figure 3 (sim2).
A: DGE detection performance of_edgeR_ applied to three different count matrices (simplesum, scaledTPM, featureCounts), with or without including an offset representing the average transcript length (for simplesum and featureCounts, avetxl indicates that such offsets were used). Including the offset or using the scaledTPM count matrix leads to improved FDR control compared to using simplesum or featureCounts matrices without offset. The curves trace out the observed FDR and TPR for each significance cutoff value. The three circles mark the performance at adjusted p-value cutoffs of 0.01, 0.05 and 0.1.B: stratification of the results inA by the presence of differential isoform usage. The improvement in FDR control seen inA results from an improved treatment of genes with differential isoform usage, while all methods perform similarly for genes without differential isoform usage.
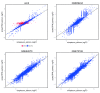
Figure 4.. Comparison of log-fold change estimates from edgeR, based on simplesum and scaledTPM count matrices, in four different data sets.
For the simulated data set (sim2), where signals have been exaggerated to pinpoint underlying causes of various observations, genes with induced DTU (whose true overall log-fold change is 0) show a clear overestimation of log-fold changes when usingsimplesum counts. However, none of the real data sets contain a similar population of genes, suggesting that for many real data sets, simple gene counting leads to overall similar conclusions as accounting for underlying changes in transcript usage.
Similar articles
- RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome.
Li B, Dewey CN. Li B, et al. BMC Bioinformatics. 2011 Aug 4;12:323. doi: 10.1186/1471-2105-12-323. BMC Bioinformatics. 2011. PMID: 21816040 Free PMC article. - Using equivalence class counts for fast and accurate testing of differential transcript usage.
Cmero M, Davidson NM, Oshlack A. Cmero M, et al. F1000Res. 2019 Mar 7;8:265. doi: 10.12688/f1000research.18276.2. eCollection 2019. F1000Res. 2019. PMID: 31143443 Free PMC article. - A robust method for transcript quantification with RNA-seq data.
Huang Y, Hu Y, Jones CD, MacLeod JN, Chiang DY, Liu Y, Prins JF, Liu J. Huang Y, et al. J Comput Biol. 2013 Mar;20(3):167-87. doi: 10.1089/cmb.2012.0230. J Comput Biol. 2013. PMID: 23461570 Free PMC article. - Comparative evaluation of full-length isoform quantification from RNA-Seq.
Sarantopoulou D, Brooks TG, Nayak S, Mrčela A, Lahens NF, Grant GR. Sarantopoulou D, et al. BMC Bioinformatics. 2021 May 25;22(1):266. doi: 10.1186/s12859-021-04198-1. BMC Bioinformatics. 2021. PMID: 34034652 Free PMC article. Review. - Computational solutions for spatial transcriptomics.
Kleino I, Frolovaitė P, Suomi T, Elo LL. Kleino I, et al. Comput Struct Biotechnol J. 2022 Sep 1;20:4870-4884. doi: 10.1016/j.csbj.2022.08.043. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36147664 Free PMC article. Review.
Cited by
- Transcriptional profiling reveals potential involvement of microvillous TRPM5-expressing cells in viral infection of the olfactory epithelium.
Baxter BD, Larson ED, Merle L, Feinstein P, Polese AG, Bubak AN, Niemeyer CS, Hassell J Jr, Shepherd D, Ramakrishnan VR, Nagel MA, Restrepo D. Baxter BD, et al. BMC Genomics. 2021 Mar 30;22(1):224. doi: 10.1186/s12864-021-07528-y. BMC Genomics. 2021. PMID: 33781205 Free PMC article. - Evolution and Expression of the Immune System of a Facultatively Anadromous Salmonid.
Colgan TJ, Moran PA, Archer LC, Wynne R, Hutton SA, McGinnity P, Reed TE. Colgan TJ, et al. Front Immunol. 2021 Feb 26;12:568729. doi: 10.3389/fimmu.2021.568729. eCollection 2021. Front Immunol. 2021. PMID: 33717060 Free PMC article. - Stromal β-catenin activation impacts nephron progenitor differentiation in the developing kidney and may contribute to Wilms tumor.
Drake KA, Chaney CP, Das A, Roy P, Kwartler CS, Rakheja D, Carroll TJ. Drake KA, et al. Development. 2020 Jul 31;147(21):dev189597. doi: 10.1242/dev.189597. Development. 2020. PMID: 32541007 Free PMC article. - TAZ-CAMTA1 and YAP-TFE3 alter the TAZ/YAP transcriptome by recruiting the ATAC histone acetyltransferase complex.
Merritt N, Garcia K, Rajendran D, Lin ZY, Zhang X, Mitchell KA, Borcherding N, Fullenkamp C, Chimenti MS, Gingras AC, Harvey KF, Tanas MR. Merritt N, et al. Elife. 2021 Apr 29;10:e62857. doi: 10.7554/eLife.62857. Elife. 2021. PMID: 33913810 Free PMC article. - Transcriptomic Signatures of Ageing Vary in Solitary and Social Forms of an Orchid Bee.
Séguret A, Stolle E, Fleites-Ayil FA, Quezada-Euán JJG, Hartfelder K, Meusemann K, Harrison MC, Soro A, Paxton RJ. Séguret A, et al. Genome Biol Evol. 2021 Jun 8;13(6):evab075. doi: 10.1093/gbe/evab075. Genome Biol Evol. 2021. PMID: 33914875 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases