FAD-I, a Fusobacterium nucleatum Cell Wall-Associated Diacylated Lipoprotein That Mediates Human Beta Defensin 2 Induction through Toll-Like Receptor-1/2 (TLR-1/2) and TLR-2/6 - PubMed (original) (raw)
FAD-I, a Fusobacterium nucleatum Cell Wall-Associated Diacylated Lipoprotein That Mediates Human Beta Defensin 2 Induction through Toll-Like Receptor-1/2 (TLR-1/2) and TLR-2/6
Sanghamitra Bhattacharyya et al. Infect Immun. 2016.
Abstract
We previously identified a cell wall-associated protein from Fusobacterium nucleatum, a Gram-negative bacterium of the oral cavity, that induces human beta defensin 2 (hBD-2) in primary human oral epithelial cells (HOECs) and designated it FAD-I (Fusobacterium-associated defensin inducer). Here, we report differential induction of hBD-2 by different strains of F. nucleatum; ATCC 25586 and ATCC 23726 induce significantly more hBD-2 mRNA than ATCC 10953. Heterologous expression of plasmid-borne fadI from the highly hBD-2-inducing strains in a ΔfadI mutant of ATCC 10953 resulted in hBD-2 induction to levels comparable to those of the highly inducing strains, indicating that FAD-I is the principal F. nucleatum agent for hBD-2 induction in HOECs. Moreover, anti-FAD-I antibodies blocked F. nucleatum induction of hBD-2 by more than 80%. Recombinant FAD-I (rFAD-I) expressed in Escherichia coli triggered levels of hBD-2 transcription and peptide release in HOECs similar to those of native FAD-I (nFAD-I) isolated from F. nucleatum ATCC 25586. Tandem mass spectrometry revealed a diacylglycerol modification at the cysteine residue in position 16 for both nFAD-I and rFAD-I. Cysteine-to-alanine substitution abrogated FAD-I's ability to induce hBD-2. Finally, FAD-I activation of hBD-2 expression was mediated via both Toll-like receptor-1/2 (TLR-1/2) and TLR-2/6 heterodimerization. Microbial molecules like FAD-I may be utilized in novel therapeutic ways to bolster the host innate immune response at mucosal surfaces.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
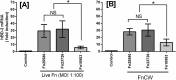
FIG 1
Induction of hBD-2 mRNA by live bacterial cells (A) and cell wall preparations (10 μg/ml cell wall F. nucleatum [Fn] ATCC 25586, ATCC 23726, and ATCC 10953 (B). The data presented are means ± standard deviations (SD) of the results of three to six replicate experiments; *, significant (P < 0.05), and NS, not significant between the indicated experiments.
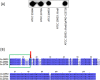
FIG 2
(A) Western immunoblot of FAD-I protein in whole-cell lysates of F. nucleatum ATCC 25586, ATCC 23726, ATCC 10953, ATCC 10953 Δ_fadI_, and ATCC 10953 Δ_fadI_/pFAD-I 23726 (ATCC 10953 Δ_fadI_ with plasmid-based expression of FAD-I from ATCC 23726). For Western blotting, whole-cell lysates derived from 1.5 × 107 cells of the different Fusobacterium strains at the mid-logarithmic growth phase were resolved in a 4 to 12% SDS-PAGE precast gel (NuPAGE, Invitrogen, USA) according to standard protocols, and anti-FAD-I antibody (1) was used to detect FAD-I. (B) Multiple-sequence alignment of FAD-I peptides from three F. nucleatum strains (ATCC 25586, ATCC 23726, and ATCC 10953). The Clustal W (54) Web-based service was used to align the sequences. The arrow indicates a conserved cysteine; the box at the N terminus indicates the signal peptide sequences; the dark-blue-shaded amino acids are common to all three proteins, while the amino acids shaded light blue or white indicate differences in sequences.
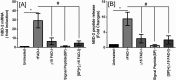
FIG 3
Induction of hBD-2 mRNA by cell wall preparations (10 μg/ml) of parent ATCC 10953, ATCC 10953 Δ_fadI_, ATCC 10953 Δ_fadI_/pFAD-I25586 (ATCC 10953 Δ_fadI_ with plasmid-based expression of FAD-I from ATCC 25586), and ATCC 10953 Δ_fadI_/pFAD-I23726 (ATCC 10953 Δ_fadI_ with plasmid-based expression of FAD-I from ATCC 23726) (A) and 25586 FnCW (10 μg/ml) (B) in the presence of anti-FAD-I antibody (1) or isotype control. The data presented are means ± SD of the results of three to six replicate experiments; *, significant (P < 0.05), and NS, not significant between the indicated treatments.
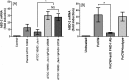
FIG 4
Comparison of hBD-2 induction and release from HOECs by rFAD-I, mature rFAD-I, and its signal peptide. HOECs were treated for 18 h with 10 μg/ml of the indicated peptides {rFAD-I, recombinant full-length FAD-I; Δ15 FAD-I, rFAD-I with the 15-amino-acid signal sequence deleted; Signal Peptide (SP), signal peptide only; [SP]+[Δ15 FAD-I], mixture of signal peptide and Δ15 FAD-I}. Fold changes in hBD-2 mRNA (A) and released peptide (B) compared to untreated HOECs were measured. The data presented are means ± SD of the results of six independent experiments; *, significant (P < 0.05) compared to untreated HOECs; #, significant (P < 0.05) compared to rFAD-I-treated cells.
FIG 5
(A and B) Tandem-mass-spectrometry chromatograms for identification of the lipid moiety in rFAD-I (A) and nFAD-I (B). The chromatogram shows the doubly charged peptide CANIDTGVDESK at m/z 914.53, where Cys modified by 576.4 Da corresponds to _S_-diacylglycerol cysteine. The asterisk indicates modification. (C and D) HOECs were treated for 18 h with 10 μg/ml of each of the peptides, as indicated [rFAD-I, recombinant FAD-I; rFAD-I (C16A), rFAD-I with the cysteine at position 16 mutated to alanine]. Fold changes in hBD-2 mRNA (C) and released peptide (D) compared to untreated HOECs were determined. The data presented are means ± SD of the results of six independent experiments; *, P < 0.05.
FIG 6
HOEC TLR-1/2 and TLR-2/6 interaction with rFAD-I. (A) HOECs were treated with either rFAD-I (10 μg/ml), Pam2Cys (a positive control for TLR-2/6; 20 ng/ml; Invivogen, USA), or Pam3Cys (a positive control for TLR-1/2; 20 ng/ml; Invivogen, USA) for 18 h in the presence or absence of anti-TLR antibodies (Invivogen, USA) as indicated. The antibodies were incubated with HOECs for 60 min prior to 18 h of incubation with the other reagents. HBD-2 mRNA fold changes compared to untreated HOECs were determined. The data presented are means ± SD of the results of three independent experiments; *, P < 0.05. (B and C) Flow cytometric analysis of TLR interaction with rFAD-I. HOECs were plated in 24-well clusters in duplicate. Upon attaining 70 to 80% confluence, the cells were harvested (0 h); left untreated (untreated); or incubated with either recombinant FAD-I (rFAD-I), Pam2Cys, or Pam3Cys for 30 min. Cells were harvested for surface staining (B) or intracellular detection (C) of TLRs by flow cytometry. (D to F) Flow cytometric analysis of TLR2 after rFAD-I and FITC-labeled rFAD-I challenge of cytochalasin D-treated HOECs. (D) Semiconfluent HOECs were harvested (0 h) and left untreated or treated with either dimethyl sulfoxide (DMSO), rFAD-I, rFAD-I plus CytD (Sigma, USA), FITC-labeled rFAD-I, or FITC-labeled rFAD-I plus CytD for 45 min. Cells were then harvested for TLR-2 surface staining and analyzed by flow cytometry. (E and F) The percentage of surface TLR2 (only)-expressing cells (E) or the percentage of surface TLR2+ rFAD-I–FITC double-positive cells (F) among HOECs harvested at 0 h, incubated with DMSO (DMSO), left untreated (untreated), or incubated with FITC-labeled rFAD-I [rFAD-I (FITC)] for 30 min in the presence or absence of 40 μM CytD was determined. The flow cytometric data presented are representative of two independent experiments.
Similar articles
- Fusobacterium nucleatum-associated beta-defensin inducer (FAD-I): identification, isolation, and functional evaluation.
Gupta S, Ghosh SK, Scott ME, Bainbridge B, Jiang B, Lamont RJ, McCormick TS, Weinberg A. Gupta S, et al. J Biol Chem. 2010 Nov 19;285(47):36523-31. doi: 10.1074/jbc.M110.133140. Epub 2010 Sep 16. J Biol Chem. 2010. PMID: 20847052 Free PMC article. - Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier.
Krisanaprakornkit S, Kimball JR, Weinberg A, Darveau RP, Bainbridge BW, Dale BA. Krisanaprakornkit S, et al. Infect Immun. 2000 May;68(5):2907-15. doi: 10.1128/IAI.68.5.2907-2915.2000. Infect Immun. 2000. PMID: 10768988 Free PMC article. - Toll-like receptor 2 and NALP2 mediate induction of human beta-defensins by fusobacterium nucleatum in gingival epithelial cells.
Ji S, Shin JE, Kim YS, Oh JE, Min BM, Choi Y. Ji S, et al. Infect Immun. 2009 Mar;77(3):1044-52. doi: 10.1128/IAI.00449-08. Epub 2008 Dec 22. Infect Immun. 2009. PMID: 19103770 Free PMC article. - Pathogenic Mechanisms of Fusobacterium nucleatum on Oral Epithelial Cells.
Groeger S, Zhou Y, Ruf S, Meyle J. Groeger S, et al. Front Oral Health. 2022 Apr 5;3:831607. doi: 10.3389/froh.2022.831607. eCollection 2022. Front Oral Health. 2022. PMID: 35478496 Free PMC article. Review.
Cited by
- Fusobacterium nucleatum - symbiont, opportunist and oncobacterium.
Brennan CA, Garrett WS. Brennan CA, et al. Nat Rev Microbiol. 2019 Mar;17(3):156-166. doi: 10.1038/s41579-018-0129-6. Nat Rev Microbiol. 2019. PMID: 30546113 Free PMC article. Review. - Use of CRISPR interference for efficient and rapid gene inactivation in Fusobacterium nucleatum.
Zhou P, G C B, Stolte F, Wu C. Zhou P, et al. Appl Environ Microbiol. 2024 Feb 21;90(2):e0166523. doi: 10.1128/aem.01665-23. Epub 2024 Jan 8. Appl Environ Microbiol. 2024. PMID: 38185820 Free PMC article. - Conceptual Perspectives: Bacterial Antimicrobial Peptide Induction as a Novel Strategy for Symbiosis with the Human Host.
Ghosh SK, Feng Z, Fujioka H, Lux R, McCormick TS, Weinberg A. Ghosh SK, et al. Front Microbiol. 2018 Feb 26;9:302. doi: 10.3389/fmicb.2018.00302. eCollection 2018. Front Microbiol. 2018. PMID: 29535688 Free PMC article. - Periodontopathogens Porphyromonas gingivalis and Fusobacterium nucleatum and Their Roles in the Progression of Respiratory Diseases.
Shi T, Wang J, Dong J, Hu P, Guo Q. Shi T, et al. Pathogens. 2023 Aug 30;12(9):1110. doi: 10.3390/pathogens12091110. Pathogens. 2023. PMID: 37764918 Free PMC article. Review. - A chemical and biological toolbox for Type Vd secretion: Characterization of the phospholipase A1 autotransporter FplA from Fusobacterium nucleatum.
Casasanta MA, Yoo CC, Smith HB, Duncan AJ, Cochrane K, Varano AC, Allen-Vercoe E, Slade DJ. Casasanta MA, et al. J Biol Chem. 2017 Dec 8;292(49):20240-20254. doi: 10.1074/jbc.M117.819144. Epub 2017 Oct 11. J Biol Chem. 2017. PMID: 29021252 Free PMC article.
References
- Krisanaprakornkit S, Kimball JR, Weinberg A, Darveau RP, Bainbridge BW, Dale BA. 2000. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect Immun 68:2907–2915. doi:10.1128/IAI.68.5.2907-2915.2000. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources