High resolution single particle refinement in EMAN2.1 - PubMed (original) (raw)
High resolution single particle refinement in EMAN2.1
James M Bell et al. Methods. 2016.
Abstract
EMAN2.1 is a complete image processing suite for quantitative analysis of grayscale images, with a primary focus on transmission electron microscopy, with complete workflows for performing high resolution single particle reconstruction, 2-D and 3-D heterogeneity analysis, random conical tilt reconstruction and subtomogram averaging, among other tasks. In this manuscript we provide the first detailed description of the high resolution single particle analysis pipeline and the philosophy behind its approach to the reconstruction problem. High resolution refinement is a fully automated process, and involves an advanced set of heuristics to select optimal algorithms for each specific refinement task. A gold standard FSC is produced automatically as part of refinement, providing a robust resolution estimate for the final map, and this is used to optimally filter the final CTF phase and amplitude corrected structure. Additional methods are in-place to reduce model bias during refinement, and to permit cross-validation using other computational methods.
Keywords: 3-D reconstruction; CryoEM; Image processing; Single particle analysis; Structural biology.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
Figure 1
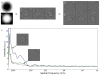
Per particle SSNR estimation. A. Exterior mask. B. Central mask. C. Raw unmasked particles. D. Background region. E. Particle region. F. Power spectra for each masked particle stack compared to traditional background estimate (dotted line), which underestimates noise and overestimates signal. SSNR is computed from the two masked curves. In this Beta-galactosidase example, there is only a modest difference between the two background calculations. In specimens with detergent or continuous carbon, the impact on SSNR estimation can be as much as an order of magnitude[33].
Figure 2
An overview of the iterative processing strategy implemented for reference-free class-averaging in e2refine2d.
Figure 3
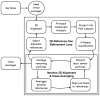
Reference-free class-averages used to produce an initial model using Monte-Carlo method implemented in e2initialmodel. Two of the possible 3-D starting maps are shown on the left along with corresponding class-average projection pairs for comparison on the right. For a good starting model, projections (1st and 3rd columns) and class-averages (2nd and 4th columns) should agree very well. The lower map exhibits poor agreement, so the higher ranked upper map would be used for 3-D refinement.
Figure 4
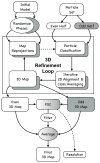
Overview of automatic refinement process described in the text and implemented in e2refine_easy. The process begins by splitting a user specified set of particles into even (white) and odd (grey) sets. Following the “gold standard” protocol, the initial model is phase randomized twice at resolutions higher than ~1.5x the target resolution. The two perturbed starting maps are then refined independently against the even and odd halves of the data. Iterative refinement begins by reprojecting the initial map and classifying particles according to their similarity to these projections. Classified particles are then iteratively aligned and averaged as shown in Fig. 2 (lower). The resulting class averages are then reconstructed in Fourier space to form a new 3D map, which becomes the starting map for the next iterative cycle. At the end of a user-specified number of iterations (typically 3–5), the process terminates. A Fourier shell correlation is computed between all pairs of maps produced from refining the even and odd subsets to assess resolution at each iteration and monitor convergence. In the final step, the even and odd maps are averaged, CTF amplitudes are corrected and the FSC is used to create a Wiener filter, ensuring that only the consistent portions of the separate refinements are visualized in the final averaged map.
Figure 5
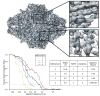
Refinement results of the Beta-galactosidase test data subset from the EMAN2.1 tutorial. e2refine_easy was run 4 times sequentially in this test, and the final FSC curves from each run are combined in one plot. The first 2 runs used downsampled data for speed, so the FSC curves do not extend to as high a resolution. The inset shows that beta-strands can be clearly resolved and alpha helices have appropriate shape. Equivalent results could have been achieved in a single run, but the intermediate results are useful in the context of the tutorial, and require less compute time. The table describes the basic parameters and wall-clock time of each refinement run. The final run was performed on a Linux cluster using 96 cores (~250 CPU-hr).
Similar articles
- New software tools in EMAN2 inspired by EMDatabank map challenge.
Bell JM, Chen M, Durmaz T, Fluty AC, Ludtke SJ. Bell JM, et al. J Struct Biol. 2018 Nov;204(2):283-290. doi: 10.1016/j.jsb.2018.09.002. Epub 2018 Sep 4. J Struct Biol. 2018. PMID: 30189321 Free PMC article. - Single-Particle Refinement and Variability Analysis in EMAN2.1.
Ludtke SJ. Ludtke SJ. Methods Enzymol. 2016;579:159-89. doi: 10.1016/bs.mie.2016.05.001. Epub 2016 Jul 1. Methods Enzymol. 2016. PMID: 27572727 Free PMC article. Review. - Single particle tomography in EMAN2.
Galaz-Montoya JG, Flanagan J, Schmid MF, Ludtke SJ. Galaz-Montoya JG, et al. J Struct Biol. 2015 Jun;190(3):279-90. doi: 10.1016/j.jsb.2015.04.016. Epub 2015 May 5. J Struct Biol. 2015. PMID: 25956334 Free PMC article. - Testing the Validity of Single-Particle Maps at Low and High Resolution.
Rosenthal PB. Rosenthal PB. Methods Enzymol. 2016;579:227-53. doi: 10.1016/bs.mie.2016.06.004. Epub 2016 Aug 8. Methods Enzymol. 2016. PMID: 27572729 Review. - Alignment algorithms and per-particle CTF correction for single particle cryo-electron tomography.
Galaz-Montoya JG, Hecksel CW, Baldwin PR, Wang E, Weaver SC, Schmid MF, Ludtke SJ, Chiu W. Galaz-Montoya JG, et al. J Struct Biol. 2016 Jun;194(3):383-94. doi: 10.1016/j.jsb.2016.03.018. Epub 2016 Mar 22. J Struct Biol. 2016. PMID: 27016284 Free PMC article.
Cited by
- Improving resolution and resolvability of single-particle cryoEM structures using Gaussian mixture models.
Chen M, Schmid MF, Chiu W. Chen M, et al. Nat Methods. 2024 Jan;21(1):37-40. doi: 10.1038/s41592-023-02082-9. Epub 2023 Nov 16. Nat Methods. 2024. PMID: 37973972 Free PMC article. - Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Jette CA, Cohen AA, Gnanapragasam PNP, Muecksch F, Lee YE, Huey-Tubman KE, Schmidt F, Hatziioannou T, Bieniasz PD, Nussenzweig MC, West AP Jr, Keeffe JR, Bjorkman PJ, Barnes CO. Jette CA, et al. bioRxiv [Preprint]. 2021 Apr 26:2021.04.23.441195. doi: 10.1101/2021.04.23.441195. bioRxiv. 2021. PMID: 33948592 Free PMC article. Updated. Preprint. - Activation mechanism and novel binding sites of the BKCa channel activator CTIBD.
Lee N, Kim S, Lee NY, Jo H, Jeong P, Pagire HS, Pagire SH, Ahn JH, Jin MS, Park CS. Lee N, et al. Life Sci Alliance. 2024 Aug 1;7(10):e202402621. doi: 10.26508/lsa.202402621. Print 2024 Oct. Life Sci Alliance. 2024. PMID: 39089879 Free PMC article. - Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Saville JW, Mannar D, Zhu X, Srivastava SS, Berezuk AM, Demers JP, Zhou S, Tuttle KS, Sekirov I, Kim A, Li W, Dimitrov DS, Subramaniam S. Saville JW, et al. Nat Commun. 2022 Feb 8;13(1):742. doi: 10.1038/s41467-022-28324-6. Nat Commun. 2022. PMID: 35136050 Free PMC article. - Multiple nanocages of a cyanophage small heat shock protein with icosahedral and octahedral symmetries.
Biswas S, Garg P, Dutta S, Suguna K. Biswas S, et al. Sci Rep. 2021 Oct 25;11(1):21023. doi: 10.1038/s41598-021-00172-2. Sci Rep. 2021. PMID: 34697325 Free PMC article.
References
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. J Struct Biol. 1996;116:190–9. - PubMed
- Frank J. Ultramicroscopy. 1981;6:343–357. - PubMed
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. J Struct Biol. 1996;116:17–24. - PubMed
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. J Struct Biol. 2007;157:38–46. - PubMed
- Hohn M, Tang G, Goodyear G, Baldwin PR, Huang Z, Penczek PA, Yang C, Glaeser RM, Adams PD, Ludtke SJ. J Struct Biol. 2007;157:47–55. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources