Ubiquitin signaling in immune responses - PubMed (original) (raw)
Review
Ubiquitin signaling in immune responses
Hongbo Hu et al. Cell Res. 2016 Apr.
Abstract
Ubiquitination has emerged as a crucial mechanism that regulates signal transduction in diverse biological processes, including different aspects of immune functions. Ubiquitination regulates pattern-recognition receptor signaling that mediates both innate immune responses and dendritic cell maturation required for initiation of adaptive immune responses. Ubiquitination also regulates the development, activation, and differentiation of T cells, thereby maintaining efficient adaptive immune responses to pathogens and immunological tolerance to self-tissues. Like phosphorylation, ubiquitination is a reversible reaction tightly controlled by the opposing actions of ubiquitin ligases and deubiquitinases. Deregulated ubiquitination events are associated with immunological disorders, including autoimmune and inflammatory diseases.
Figures
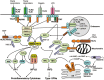
Figure 1
Main signaling pathways of PRRs. A common signaling feature of different PRRs is the dependence on adaptor molecules. TLRs, except TLR3, associate with MyD88, which in turn recruits and activates IRAK4, leading to phosphorylation of IRAK1 and IRAK1-mediated activation of the downstream adaptor TRAF6. TLR3 and TLR4 associate with TRIF and activate the downstream adaptor RIP1. Via a ubiquitin-dependent mechanism, TRAF6 and RIP1 both activate TAK1 and its downstream pathways, leading to induction of proinflammatory cytokines. TRIF also activates TBK1 (or IKKε) for the induction of type I IFNs, via a mechanism thought to involve the adaptor TRAF3. RLRs signal via the mitochondrial adaptor MAVS and several TRAF members to activate the TAK1 proinflammatory pathway and the type I IFN pathway, whereas the NLR member NOD2 (as well as NOD1) activates the TAK1 proinflammatory pathway via the signaling adaptor RIP2. STING is a signaling adaptor of cytoplasmic DNA sensors, such as cGAS, which activates STING via synthesizing the second messenger, cyclic dinucleotide cGAMP. STING also senses bacteria-derived cyclic di-nucleotides, c-di-AMP and c-di-GMP.
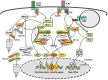
Figure 2
Ubiquitin regulation of TLR signaling. In response to MyD88- and TRIF-dependent TLR signals, TRAF6 undergoes self-ubiquitination and RIP1 is ubiquitinated by Peli1, an E3 ligase that is activated via its phosphorylation by IRAK1 and TBK1 or IKKε. It is generally thought that K63-linked ubiquitin chains conjugated to TRAF6 and RIP1 facilitate the recruitment and activation of TAK1 and IKK, which requires the ubiquitin-binding proteins TAB2 (or TAB3) and NEMO. Activated IKK and MAPKs (downstream of TAK1) mediate activation of NF-κB c-Rel and RelA, and members of the AP1 family, which together with TRAF6-stimulated IRF5 transactivate a variety of proinflammatory cytokine genes. As discussed in the text, this signaling model requires further modifications, since some parts are not supported by knockout mouse studies. TLR signaling is subject to control by negative regulators, one of which is TRAF3 that negatively regulates TLR-stimulated proinflammatory signaling by two possible mechanisms: interfering with the signaling function of the MyD88 complex and mediating degradation of IRF5 and c-Rel by forming an E3 complex with TRAF2 and cIAP. At least in the TLR4 signaling pathway, TRAF3 is targeted for ubiquitin-dependent degradation via a mechanism in which Peli1 either cooperates with TRAF6 or functions downstream of TRAF6 to mediate K63-linked ubiquitination and activation of cIAP, allowing the activated cIAP to conjugate K48-linked ubiquitin chains to TRAF3. By mediating TRAF3 degradation, Peli1 promotes proinflammatory TLR signaling.
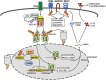
Figure 3
Ubiquitin regulation of DC activation. Dendritic cells (DCs) are primary antigen-presenting cells (APCs) that are activated by signaling from PRRs, such as TLRs, during an infection. Activated DCs undergo maturation, including upregulation of MHCII and the costimulatory molecule CD86 that are required for antigen presentation and T cell costimulation. Activated DCs also produce various cytokines, such as the proinflammatory cytokines IL-12 and IL-23, which in turn regulate the differentiation of antigen-activated T cells. The membrane-bound E3 ubiquitin ligase MARCH1 mediates ubiquitin-dependent internalization and lysosomal degradation of MHCII and CD86, thereby negatively regulating the function of DCs in T cell activation. The rhomboid family protease Rhbdd3 negatively regulates DC activation by binding to NEMO and recruiting the DUB A20 for deconjugation of K63-linked ubiquitin chains from NEMO. Trabid is a member of the A20 subfamily of OTU-containing DUBs that deubiquitinates and stabilizes Jmjd2d, a histone demethylase that removes transcriptionally repressive histone modifications, H3K9me2 and H3K9me3, from the Il12 (Il12a and Il12b) and Il23 (Il23a) promoters to promote the recruitment of c-Rel-containing NF-κB complexes to the κB enhancer element for Il12/Il23 transcription. The E3 ubiquitin ligase catalyzing Jmjd2d ubiquitination is currently unknown.
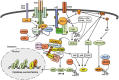
Figure 4
Ubiquitin regulation of T cell activation. TCR signaling is initiated by antigen/MHC-stimulated colocalization of the TCR complex with T cell coreceptor, CD4 or CD8, which allows the CD4/CD8-associated protein tyrosine kinase Lck to phosphorylate the signaling chains of TCR. Phosphorylated TCRζ chains recruit and activate the protein tyrosine kinase Zap70, which in turn phosphorylates LAT and SLP-76, triggering the recruitment and activation of key signaling components, such as PLCγ1. PLCγ1 hydrolyzes the membrane lipid PIP2 to generate second messengers, DAG and IP3, which in turn mediate activation of PKCθ, Ras, and calcium pathways, leading to activation of NF-κB, AP1, and NFAT. K63-linked ubiquitination is crucial for activation of NF-κB and AP1 pathways by the intermediate signaling complex, composed of CARMA1, BCL10, and MALT1. This complex associates with an E3 and the E2 dimer Ubc13/Uev1a to conjugate K63-linked ubiquitin chains to BCL10, thereby promoting the recruitment and activation of TAK1 and IKK. Several E3 ubiquitin ligases, including Cbl-b, c-Cbl, GRAIL, and ITCH, negatively regulate TCR-proximal signaling by mediating ubiquitin-dependent degradation of major signaling components. Some E3 ligases, likely including Nrdp1 and c-Cbl, conjugate nondegradative ubiquitin chains, including K33-linked chains, to Zap70, which facilitates the recruitment of protein tyrosine phosphatases Sts1 and Sts2 to dephosphorylate and inhibit Zap70. Peli1 and MDM2 are E3s that mediate ubiquitin-dependent degradation of activated transcription factors, c-Rel and NFATc2, respectively.
Similar articles
- Ubiquitylation in innate and adaptive immunity.
Bhoj VG, Chen ZJ. Bhoj VG, et al. Nature. 2009 Mar 26;458(7237):430-7. doi: 10.1038/nature07959. Nature. 2009. PMID: 19325622 Review. - Ubiquitin Ligases in Cancer Immunotherapy - Balancing Antitumor and Autoimmunity.
Fujita Y, Tinoco R, Li Y, Senft D, Ronai ZA. Fujita Y, et al. Trends Mol Med. 2019 May;25(5):428-443. doi: 10.1016/j.molmed.2019.02.002. Epub 2019 Mar 18. Trends Mol Med. 2019. PMID: 30898473 Free PMC article. Review. - Role of Ubiquitin Signaling in Modulating Dendritic Cell Function.
Jie Z. Jie Z. Adv Exp Med Biol. 2024;1466:101-111. doi: 10.1007/978-981-97-7288-9_7. Adv Exp Med Biol. 2024. PMID: 39546138 Review. - To TRIM the Immunity: From Innate to Adaptive Immunity.
Yang W, Gu Z, Zhang H, Hu H. Yang W, et al. Front Immunol. 2020 Oct 8;11:02157. doi: 10.3389/fimmu.2020.02157. eCollection 2020. Front Immunol. 2020. PMID: 33117334 Free PMC article. - Deubiquitinases as pivotal regulators of T cell functions.
Yang XD, Sun SC. Yang XD, et al. Front Med. 2018 Aug;12(4):451-462. doi: 10.1007/s11684-018-0651-y. Epub 2018 Jul 27. Front Med. 2018. PMID: 30054854 Free PMC article. Review.
Cited by
- Fucosylated ubiquitin and orthogonally glycosylated mutant A28C: conceptually new ligands for Burkholderia ambifaria lectin (BambL).
Kuhaudomlarp S, Cerofolini L, Santarsia S, Gillon E, Fallarini S, Lombardi G, Denis M, Giuntini S, Valori C, Fragai M, Imberty A, Dondoni A, Nativi C. Kuhaudomlarp S, et al. Chem Sci. 2020 Oct 21;11(47):12662-12670. doi: 10.1039/d0sc03741a. Chem Sci. 2020. PMID: 34094460 Free PMC article. - Use of Extracellular Monomeric Ubiquitin as a Therapeutic Option for Major Depressive Disorder.
Maldonado-García JL, García-Mena LH, Mendieta-Cabrera D, Pérez-Sánchez G, Becerril-Villanueva E, Alvarez-Herrera S, Homberg T, Vallejo-Castillo L, Pérez-Tapia SM, Moreno-Lafont MC, Ortuño-Sahagún D, Pavón L. Maldonado-García JL, et al. Pharmaceuticals (Basel). 2024 Jun 27;17(7):841. doi: 10.3390/ph17070841. Pharmaceuticals (Basel). 2024. PMID: 39065692 Free PMC article. - Cytosolic DNA sensing by cGAS: regulation, function, and human diseases.
Yu L, Liu P. Yu L, et al. Signal Transduct Target Ther. 2021 Apr 30;6(1):170. doi: 10.1038/s41392-021-00554-y. Signal Transduct Target Ther. 2021. PMID: 33927185 Free PMC article. Review. - Ubiquitin Proteasome Gene Signatures in Ependymoma Molecular Subtypes.
Vriend J, Thanasupawat T, Sinha N, Klonisch T. Vriend J, et al. Int J Mol Sci. 2022 Oct 15;23(20):12330. doi: 10.3390/ijms232012330. Int J Mol Sci. 2022. PMID: 36293188 Free PMC article. Review. - Novel candidate genes for ECT response prediction-a pilot study analyzing the DNA methylome of depressed patients receiving electroconvulsive therapy.
Moschny N, Zindler T, Jahn K, Dorda M, Davenport CF, Wiehlmann L, Maier HB, Eberle F, Bleich S, Neyazi A, Frieling H. Moschny N, et al. Clin Epigenetics. 2020 Jul 29;12(1):114. doi: 10.1186/s13148-020-00891-9. Clin Epigenetics. 2020. PMID: 32727556 Free PMC article.
References
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998; 67:425–479. - PubMed
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem 2009; 78:399–434. - PubMed
- Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol 2014; 21:301–307. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI064639/AI/NIAID NIH HHS/United States
- AI104519/AI/NIAID NIH HHS/United States
- AI064639/AI/NIAID NIH HHS/United States
- R01 AI057555/AI/NIAID NIH HHS/United States
- GM84459/GM/NIGMS NIH HHS/United States
- R37 AI064639/AI/NIAID NIH HHS/United States
- R01 AI104519/AI/NIAID NIH HHS/United States
- AI057555/AI/NIAID NIH HHS/United States
- R01 GM084459/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources