Alignment algorithms and per-particle CTF correction for single particle cryo-electron tomography - PubMed (original) (raw)
Alignment algorithms and per-particle CTF correction for single particle cryo-electron tomography
Jesús G Galaz-Montoya et al. J Struct Biol. 2016 Jun.
Abstract
Single particle cryo-electron tomography (cryoSPT) extracts features from cryo-electron tomograms, followed by 3D classification, alignment and averaging to generate improved 3D density maps of such features. Robust methods to correct for the contrast transfer function (CTF) of the electron microscope are necessary for cryoSPT to reach its resolution potential. Many factors can make CTF correction for cryoSPT challenging, such as lack of eucentricity of the specimen stage, inherent low dose per image, specimen charging, beam-induced specimen motions, and defocus gradients resulting both from specimen tilting and from unpredictable ice thickness variations. Current CTF correction methods for cryoET make at least one of the following assumptions: that the defocus at the center of the image is the same across the images of a tiltseries, that the particles all lie at the same Z-height in the embedding ice, and/or that the specimen, the cryo-electron microscopy (cryoEM) grid and/or the carbon support are flat. These experimental conditions are not always met. We have developed a CTF correction algorithm for cryoSPT without making any of the aforementioned assumptions. We also introduce speed and accuracy improvements and a higher degree of automation to the subtomogram averaging algorithms available in EMAN2. Using motion-corrected images of isolated virus particles as a benchmark specimen, recorded with a DE20 direct detection camera, we show that our CTF correction and subtomogram alignment routines can yield subtomogram averages close to 4/5 Nyquist frequency of the detector under our experimental conditions.
Keywords: Contrast transfer function (CTF); Cryo-electron tomography (cryoET); Direct detection device (DDD); EMAN2; Single particle cryo-electron tomography (cryoSPT); Subtomogram averaging.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
Figure 1. Drift correction for cryoET images of a tilted specimen collected with a DE20 camera
Examples of VEEV images exhibiting significant drift, taken at different tilt angles, shown before (top) and after (bottom) drift correction.
Figure 2. Quantification of image drift in cryoET images of a tilted specimen
Examples of total drift per frame and average drift per image for all images in (A) a “bad” unidirectional tiltseries and (B) a “good” bidirectional tiltseries.
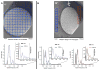
Figure 3. Interactive CTF fitting in EMAN2
Examples of periodogram averaging by interactive micrograph tiling for power spectrum (PS) computation and CTF fitting directly from the imaging area for cryoET images of a tilted specimen. (A) Tiling of 0° image from a VEEV tiltseries showing clear CTF ripples out to 4/5 Nyquist. (B) Example of per-strip tiling of a high tilt (50°) image from a VEEV tiltseries, showing clear CTF ripples out to 2/3 Nyquist.
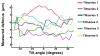
Figure 4. Measured defocus variation within and across tiltseries
Plots showing large measured defocus variations within and across six VEEV cryoET tiltseries collected using a JEM3200FSC electron microscope.
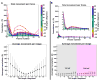
Figure 5. Resolution analyses of CTF-Corrected VEEV subtomogram averages
(A) Per-tomogram FSC curves for six CTF-corrected VEEV subtomogram averages from independent tomograms keeping the top 75% best correlating particles in each average. (B) FSCs for CTF-corrected subtomogram averages comprised of different fractions of particles (~100% / 516 particles; ~90% / 464 particles; ~75% / 387 particles; ~30% / 154 particles; ~10% / 51 particles).
Figure 6. CTF-Corrected Subtomogram Average of VEEV
Isosurfaces of VEEV subtomogram average comprised of the top 75% best-correlating subtomograms showing the outer glycoprotein shell (top) and the nucleocapsid (bottom).
Similar articles
- Efficient 3D-CTF correction for cryo-electron tomography using NovaCTF improves subtomogram averaging resolution to 3.4Å.
Turoňová B, Schur FKM, Wan W, Briggs JAG. Turoňová B, et al. J Struct Biol. 2017 Sep;199(3):187-195. doi: 10.1016/j.jsb.2017.07.007. Epub 2017 Jul 22. J Struct Biol. 2017. PMID: 28743638 Free PMC article. - TomoAlign: A novel approach to correcting sample motion and 3D CTF in CryoET.
Fernandez JJ, Li S. Fernandez JJ, et al. J Struct Biol. 2021 Dec;213(4):107778. doi: 10.1016/j.jsb.2021.107778. Epub 2021 Aug 18. J Struct Biol. 2021. PMID: 34416376 - Quantifying resolution limiting factors in subtomogram averaged cryo-electron tomography using simulations.
Voortman LM, Vulović M, Maletta M, Voigt A, Franken EM, Simonetti A, Peters PJ, van Vliet LJ, Rieger B. Voortman LM, et al. J Struct Biol. 2014 Aug;187(2):103-111. doi: 10.1016/j.jsb.2014.06.007. Epub 2014 Jul 3. J Struct Biol. 2014. PMID: 24998892 - Cryo-Electron Tomography and Subtomogram Averaging.
Wan W, Briggs JA. Wan W, et al. Methods Enzymol. 2016;579:329-67. doi: 10.1016/bs.mie.2016.04.014. Epub 2016 Jun 22. Methods Enzymol. 2016. PMID: 27572733 Review. - Subtomogram averaging from cryo-electron tomograms.
Leigh KE, Navarro PP, Scaramuzza S, Chen W, Zhang Y, Castaño-Díez D, Kudryashev M. Leigh KE, et al. Methods Cell Biol. 2019;152:217-259. doi: 10.1016/bs.mcb.2019.04.003. Epub 2019 May 15. Methods Cell Biol. 2019. PMID: 31326022 Review.
Cited by
- Promotion of virus assembly and organization by the measles virus matrix protein.
Ke Z, Strauss JD, Hampton CM, Brindley MA, Dillard RS, Leon F, Lamb KM, Plemper RK, Wright ER. Ke Z, et al. Nat Commun. 2018 Apr 30;9(1):1736. doi: 10.1038/s41467-018-04058-2. Nat Commun. 2018. PMID: 29712906 Free PMC article. - Missing Wedge Completion via Unsupervised Learning with Coordinate Networks.
Van Veen D, Galaz-Montoya JG, Shen L, Baldwin P, Chaudhari AS, Lyumkis D, Schmid MF, Chiu W, Pauly J. Van Veen D, et al. Int J Mol Sci. 2024 May 17;25(10):5473. doi: 10.3390/ijms25105473. Int J Mol Sci. 2024. PMID: 38791508 Free PMC article. - Tools for visualizing and analyzing Fourier space sampling in Cryo-EM.
Baldwin PR, Lyumkis D. Baldwin PR, et al. Prog Biophys Mol Biol. 2021 Mar;160:53-65. doi: 10.1016/j.pbiomolbio.2020.06.003. Epub 2020 Jul 6. Prog Biophys Mol Biol. 2021. PMID: 32645314 Free PMC article. Review. - Fine details in complex environments: the power of cryo-electron tomography.
Hutchings J, Zanetti G. Hutchings J, et al. Biochem Soc Trans. 2018 Aug 20;46(4):807-816. doi: 10.1042/BST20170351. Epub 2018 Jun 22. Biochem Soc Trans. 2018. PMID: 29934301 Free PMC article. Review. - Neutralizing Antibodies Inhibit Chikungunya Virus Budding at the Plasma Membrane.
Jin J, Galaz-Montoya JG, Sherman MB, Sun SY, Goldsmith CS, O'Toole ET, Ackerman L, Carlson LA, Weaver SC, Chiu W, Simmons G. Jin J, et al. Cell Host Microbe. 2018 Sep 12;24(3):417-428.e5. doi: 10.1016/j.chom.2018.07.018. Epub 2018 Aug 23. Cell Host Microbe. 2018. PMID: 30146390 Free PMC article.
References
- Asano S, Fukuda Y, Beck F, Aufderheide A, Förster F, Danev R, Baumeister W. A molecular census of 26S proteasomes in intact neurons. Science. 2015;347(6220):439–442. - PubMed
Publication types
MeSH terms
Grants and funding
- P41 GM103832/GM/NIGMS NIH HHS/United States
- R01 GM080139/GM/NIGMS NIH HHS/United States
- R24 AI120942/AI/NIAID NIH HHS/United States
- U54 AI057156/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous