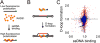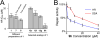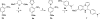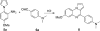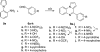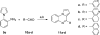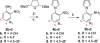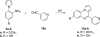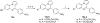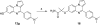Development of Small Molecules that Specifically Inhibit the D-loop Activity of RAD51 - PubMed (original) (raw)
Development of Small Molecules that Specifically Inhibit the D-loop Activity of RAD51
Wei Lv et al. J Med Chem. 2016.
Abstract
RAD51 is the central protein in homologous recombination (HR) DNA repair and represents a therapeutic target in oncology. Herein we report a novel class of RAD51 inhibitors that were identified by high throughput screening. In contrast to many previously reported RAD51 inhibitors, our lead compound 1 is capable of blocking RAD51-mediated D-loop formation (IC50 21.3 ± 7.8 μM) at concentrations that do not influence RAD51 binding to ssDNA. In human cells, 1 inhibits HR (IC50 13.1 ± 1.6 μM) without blocking RAD51's ability to assemble into subnuclear foci at sites of DNA damage. We determined that the active constituent of 1 is actually an oxidized derivative (termed RI(dl)-1 or 8) of the original screening compound. Our SAR campaign also yielded RI(dl)-2 (hereafter termed 9h), which effectively blocks RAD51's D-loop activity in biochemical systems (IC50 11.1 ± 1.3 μM) and inhibits HR activity in human cells (IC50 3.0 ± 1.8 μM).
Conflict of interest statement
Notes
The authors declare no competing financial interest.
Figures
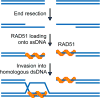
Figure 1
Schematic representation of the mechanism of DNA double-strand breaks repair by homologous recombination. Following a DSB, the DNA ends are resected to generate 3′ ssDNA overhangs onto which RAD51 loads. The resulting RAD51-ssDNA nucleoprotein filament is capable of invading homologous dsDNA and base-pairing with complementary DNA, thereby forming a heteroduplex and displacing a loop (D-loop) of ssDNA.
Figure 2
High-throughput (HT) screen for compounds that inhibit RAD51’s D-loop activity. (A) One biochemical assay monitors RAD51 binding to fluorescently labeled ssDNA, which is detected as an increase in fluorescence polarization. (B) A second parallel biochemical assay monitors fluorescence intensity, which decreases upon pairing of Black Hole Quencher 1-labeled ssDNA with a fluorescein-labeled complementary double-hairpin duplex. (C) Compounds were tested for the ability to influence the efficiency of these two RAD51-mediated processes, and the results of this HT are displayed. Blue symbols = ASDI library compounds, red symbols = LOPAC library compounds. Arrow indicates the position of compound 1.
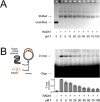
Figure 3
Compound 1 inhibits RAD51’s D-loop activity with minimal interference of RAD51-ssDNA binding. (A) Electromobility shift assay showing that 1 does not inhibit interaction with RAD51 and ssDNA. (B) Gel-based D-loop formation assay showing that 1 inhibits RAD51-mediated assimilation of a radiolabeled ssDNA oligonucleotide into the homologous region of a supercoiled dsDNA plasmid by up to 74%. Error bars indicate the standard error for four replicates with a representative gel image shown.
Figure 4
An overview is shown for the optimization strategy of 8.
Figure 5
(A) Inhibition of cellular HR activity by 1 and optimized analogues of 1 using the DR-GFP assay. Data were collected at 24 h following transfection with the I-SceI expressing plasmid pCBASce. Error bars denote the standard error for three replicates. (B) Inhibition of cellular HR and SSA activity by 9h at 24 h post-transfection. Error bars denote the standard error for three replicates.
Figure 6
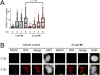
(A) Compound 9h does not inhibit the appearance of IR-induced RAD51 foci in 293-DR-GFP cells over an 8 h time course. At least 100 nuclei per condition are represented. (B) Representative micrographs showing colocalization of RAD51 foci with replication protein A (RPA) foci in DAPI-counterstained 293-DR-GFP nuclei at 8 h postirradiation in 293-DR-GFP cells treated with 9h or the DMSO vehicle control.
Figure 7
(A,B) Electromobility shift assay showing RAD51-ssDNA binding in the presence of 9h or RI-1, respectively. (C) 9h does not destabilize RAD51-ssDNA nucleoprotein filaments as shown by salt titration midpoint. RI-1 serves as a positive control for disruption of RAD51-ssDNA nucleoprotein filament stability. Error bars denote the standard error for three replicates. (D,E,F) Representative D-loop assay gel images showing recombinase-mediated assimilation of a radiolabeled ssDNA oligonucleotide into the homologous region of a supercoiled dsDNA plasmid in the presence of 9h by human RAD51 protein, S. cerevisiae Rad51 and Rad52 proteins, or E. coli RecA protein, respectively. Error bars denote the standard error for three replicates.
Figure 8
Clonogenic survival of (A) U2OS, (B) PC-3, and (C) MCF-7 tumor cell lines treated with ionizing radiation followed by outgrowth in the presence of 9h or the vehicle-only control. Error bars denote the standard error for three replicates.
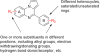
Scheme 1. Synthesis of 7_a_
_a_Reagents and conditions: (a) AcOH, reflux, 3 h, 90%; (b) H2, Pd/C, MeOH, 5 h, 97%; (c) 1,2,3-benzotriazole, AlCl3, THF, 4 h, 32%.
Scheme 2. Synthesis of 8_a_
_a_Reagents and conditions: (a) 1,2,3-benzotriazole, AlCl3, THF, overnight; (b) MnO2, acetone, 55 °C, 24 h, 59% in two steps.
Scheme 3. Synthesis of Analogues 9a–j_a_
_a_Reagents and conditions: (a) 1,2,3-benzotriazole, AlCl3, THF, overnight; (b) MnO2, acetone, 55 °C, 24 h, 19–70% in two steps; (c) H2, Pd/C, methanol, 5 h, 57–87%.
Scheme 4. Synthesis of Analogues 11a–d_a_
_a_Reagents and conditions: (a) 1,2,3-benzotriazole, AlCl3, THF, overnight; (b) MnO2, acetone, 55 °C, 24 h, 35–71% in two steps.
Scheme 5. Synthesis of 5b–d_a_
_a_Reagents and conditions: (a) AcOH, reflux, 3 h, 45–63%; (b) H2, Pd/C, MeOH, 5 h, 37–97%.
Scheme 6. Synthesis of Analogues 12a–m_a_
_a_Reagents and conditions: (a) 1,2,3-benzotriazole, AlCl3, THF, overnight; (b) MnO2, acetone, 55 °C, 24 h, 7–77% in two steps; (c) H2, Pd/C, MeOH, 5 h, 50–93%.
Scheme 7. Synthesis of Analogues 13a–b_a_
_a_Reagents and conditions: (a) 1,2,3-benzotriazole, AlCl3, THF, overnight; (b) MnO2, acetone, 55 °C, 24 h, 52–78% in two steps.
Scheme 8. Synthesis of Analogues 15a–b_a_
_a_Reagents and conditions: (a) allyl bromide, Cs2CO3, DMF, microwave, 80 °C, 30 min, 58%; (b) FCH2CH2OH, DEAD, Ph3P, THF, microwave, 60 °C, 40 min; (c) H2, Pd/C, MeOH, overnight, 36–45%.
Scheme 9. Synthesis of Analogues 16_a_
_a_Reagents and conditions: (a) Br(CH3)2CHCONH2, NaOH, DMA, 50 °C, 3 h, 41%.
Similar articles
- Recent Developments Using Small Molecules to Target RAD51: How to Best Modulate RAD51 for Anticancer Therapy?
Budke B, Lv W, Kozikowski AP, Connell PP. Budke B, et al. ChemMedChem. 2016 Nov 21;11(22):2468-2473. doi: 10.1002/cmdc.201600426. Epub 2016 Oct 26. ChemMedChem. 2016. PMID: 27781374 Free PMC article. Review. - Optimization of Drug Candidates That Inhibit the D-Loop Activity of RAD51.
Budke B, Tueckmantel W, Miles K, Kozikowski AP, Connell PP. Budke B, et al. ChemMedChem. 2019 May 17;14(10):1031-1040. doi: 10.1002/cmdc.201900075. Epub 2019 Apr 17. ChemMedChem. 2019. PMID: 30957434 Free PMC article. - Identification and characterization of human Rad51 inhibitors by screening of an existing drug library.
Normand A, Rivière E, Renodon-Cornière A. Normand A, et al. Biochem Pharmacol. 2014 Oct 1;91(3):293-300. doi: 10.1016/j.bcp.2014.07.033. Epub 2014 Aug 11. Biochem Pharmacol. 2014. PMID: 25124703 - Quinazolinone derivatives as inhibitors of homologous recombinase RAD51.
Ward A, Dong L, Harris JM, Khanna KK, Al-Ejeh F, Fairlie DP, Wiegmans AP, Liu L. Ward A, et al. Bioorg Med Chem Lett. 2017 Jul 15;27(14):3096-3100. doi: 10.1016/j.bmcl.2017.05.039. Epub 2017 May 15. Bioorg Med Chem Lett. 2017. PMID: 28545975 - Targeting homologous recombination, new pre-clinical and clinical therapeutic combinations inhibiting RAD51.
Ward A, Khanna KK, Wiegmans AP. Ward A, et al. Cancer Treat Rev. 2015 Jan;41(1):35-45. doi: 10.1016/j.ctrv.2014.10.006. Epub 2014 Nov 1. Cancer Treat Rev. 2015. PMID: 25467108 Review.
Cited by
- Exploiting DNA repair pathways for tumor sensitization, mitigation of resistance, and normal tissue protection in radiotherapy.
Nickoloff JA, Taylor L, Sharma N, Kato TA. Nickoloff JA, et al. Cancer Drug Resist. 2021;4(2):244-263. doi: 10.20517/cdr.2020.89. Epub 2021 Jun 19. Cancer Drug Resist. 2021. PMID: 34337349 Free PMC article. - Regulation and pharmacological targeting of RAD51 in cancer.
Grundy MK, Buckanovich RJ, Bernstein KA. Grundy MK, et al. NAR Cancer. 2020 Sep;2(3):zcaa024. doi: 10.1093/narcan/zcaa024. Epub 2020 Sep 25. NAR Cancer. 2020. PMID: 33015624 Free PMC article. - SARS-CoV-2 exploits cellular RAD51 to promote viral propagation: implication of RAD51 inhibitor as a potential drug candidate against COVID-19.
Pham TX, Huynh TTX, Choi J, Lee J-B, Park S-C, Kim B, Lim Y-S, Hwang SB. Pham TX, et al. J Virol. 2023 Dec 21;97(12):e0173723. doi: 10.1128/jvi.01737-23. Epub 2023 Dec 5. J Virol. 2023. PMID: 38051260 Free PMC article. - Dual PARP and RAD51 Inhibitory Drug Conjugates Show Synergistic and Selective Effects on Breast Cancer Cells.
Malka MM, Eberle J, Niedermayer K, Zlotos DP, Wiesmüller L. Malka MM, et al. Biomolecules. 2021 Jul 3;11(7):981. doi: 10.3390/biom11070981. Biomolecules. 2021. PMID: 34356606 Free PMC article. - Recent Developments Using Small Molecules to Target RAD51: How to Best Modulate RAD51 for Anticancer Therapy?
Budke B, Lv W, Kozikowski AP, Connell PP. Budke B, et al. ChemMedChem. 2016 Nov 21;11(22):2468-2473. doi: 10.1002/cmdc.201600426. Epub 2016 Oct 26. ChemMedChem. 2016. PMID: 27781374 Free PMC article. Review.
References
- Thompson LH, Schild D. Homologous Recombinational Repair of DNA Ensures Mammalian Chromosome Stability. Mutat Res, Fundam Mol Mech Mutagen. 2001;477:131–153. - PubMed
- Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, Brookman KW, Siciliano MJ, Walter CA, Fan W, Narayana LS, Zhou ZQ, Adamson AW, Sorensen KJ, Chen DJ, Jones NJ, Thompson LH. Xrcc2 and Xrcc3, New Human Rad51-Family Members, Promote Chromosome Stability and Protect against DNA Cross-Links and Other Damages. Mol Cell. 1998;1:783–793. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Research Materials
Miscellaneous