Design and computational analysis of single-cell RNA-sequencing experiments - PubMed (original) (raw)
Review
Design and computational analysis of single-cell RNA-sequencing experiments
Rhonda Bacher et al. Genome Biol. 2016.
Abstract
Single-cell RNA-sequencing (scRNA-seq) has emerged as a revolutionary tool that allows us to address scientific questions that eluded examination just a few years ago. With the advantages of scRNA-seq come computational challenges that are just beginning to be addressed. In this article, we highlight the computational methods available for the design and analysis of scRNA-seq experiments, their advantages and disadvantages in various settings, the open questions for which novel methods are needed, and expected future developments in this exciting area.
Figures
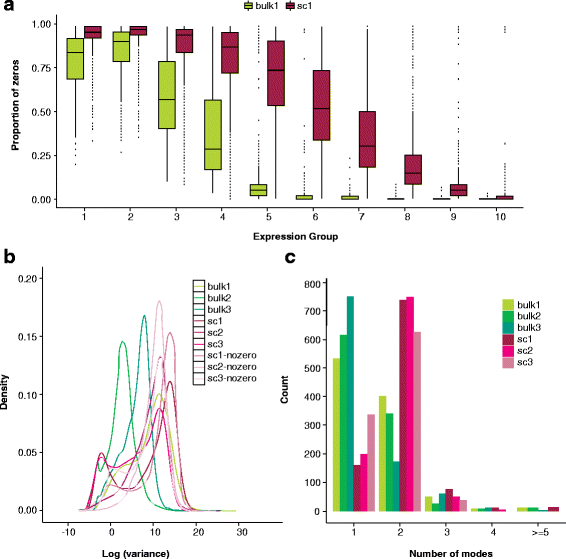
Fig. 1
Prominent features in single-cell RNA-seq data relative to bulk RNA-seq include an abundance of zeros, increased variability, and multi-modal expression distributions. a Boxplots of the gene-specific proportion of zeros in a bulk (bulk1) and single-cell (sc1) dataset stratified by percentile of median gene expression. Sequencing depth ranges from 420,000 to 16.6 million in bulk1 and 385,000 to 16.4 million in sc1 (samples were chosen to have comparable depths; see the “Data” section). b Densities of gene-specific log variance for all genes in three bulk and three single-cell RNA-seq datasets. Densities are also shown for the single-cell datasets for log variances calculated following the removal of zeros, emphasizing that the increased variability observed relative to bulk is not entirely due to the presence of zeros. c For each dataset shown in b, 1000 genes were selected at random from the list of genes for which at least 75 % of cells showed non-zero expression. For each gene, zeros were removed and Mclust [92] was applied to log expression to estimate the number of modes. Because zeros were removed prior to Mclust, a mode at zero will not contribute to the total number of modes shown
Similar articles
- Data Analysis in Single-Cell Transcriptome Sequencing.
Gao S. Gao S. Methods Mol Biol. 2018;1754:311-326. doi: 10.1007/978-1-4939-7717-8_18. Methods Mol Biol. 2018. PMID: 29536451 - Computational approaches for interpreting scRNA-seq data.
Rostom R, Svensson V, Teichmann SA, Kar G. Rostom R, et al. FEBS Lett. 2017 Aug;591(15):2213-2225. doi: 10.1002/1873-3468.12684. Epub 2017 Jun 12. FEBS Lett. 2017. PMID: 28524227 Free PMC article. Review. - An accessible, interactive GenePattern Notebook for analysis and exploration of single-cell transcriptomic data.
Mah CK, Wenzel AT, Juarez EF, Tabor T, Reich MM, Mesirov JP. Mah CK, et al. F1000Res. 2018 Aug 16;7:1306. doi: 10.12688/f1000research.15830.2. eCollection 2018. F1000Res. 2018. PMID: 31316748 Free PMC article. - Cell lineage inference from SNP and scRNA-Seq data.
Ding J, Lin C, Bar-Joseph Z. Ding J, et al. Nucleic Acids Res. 2019 Jun 4;47(10):e56. doi: 10.1093/nar/gkz146. Nucleic Acids Res. 2019. PMID: 30820578 Free PMC article. - The promise of single-cell RNA sequencing for kidney disease investigation.
Wu H, Humphreys BD. Wu H, et al. Kidney Int. 2017 Dec;92(6):1334-1342. doi: 10.1016/j.kint.2017.06.033. Epub 2017 Oct 12. Kidney Int. 2017. PMID: 28893418 Free PMC article. Review.
Cited by
- Modeling dynamic correlation in zero-inflated bivariate count data with applications to single-cell RNA sequencing data.
Yang Z, Ho YY. Yang Z, et al. Biometrics. 2022 Jun;78(2):766-776. doi: 10.1111/biom.13457. Epub 2021 Mar 30. Biometrics. 2022. PMID: 33720414 Free PMC article. - Unveiling the novel immune and molecular signatures of ovarian cancer: insights and innovations from single-cell sequencing.
Li Z, Gu H, Xu X, Tian Y, Huang X, Du Y. Li Z, et al. Front Immunol. 2023 Nov 1;14:1288027. doi: 10.3389/fimmu.2023.1288027. eCollection 2023. Front Immunol. 2023. PMID: 38022625 Free PMC article. Review. - Aortic Cellular Diversity and Quantitative Genome-Wide Association Study Trait Prioritization Through Single-Nuclear RNA Sequencing of the Aneurysmal Human Aorta.
Chou EL, Chaffin M, Simonson B, Pirruccello JP, Akkad AD, Nekoui M, Lino Cardenas CL, Bedi KC Jr, Nash C, Juric D, Stone JR, Isselbacher EM, Margulies KB, Klattenhoff C, Ellinor PT, Lindsay ME. Chou EL, et al. Arterioscler Thromb Vasc Biol. 2022 Nov;42(11):1355-1374. doi: 10.1161/ATVBAHA.122.317953. Epub 2022 Sep 29. Arterioscler Thromb Vasc Biol. 2022. PMID: 36172868 Free PMC article. - Data-driven assessment of dimension reduction quality for single-cell omics data.
Dong X, Bacher R. Dong X, et al. Patterns (N Y). 2022 Mar 11;3(3):100465. doi: 10.1016/j.patter.2022.100465. eCollection 2022 Mar 11. Patterns (N Y). 2022. PMID: 35510193 Free PMC article. - Single-cell RNA sequencing reveals gene expression signatures of breast cancer-associated endothelial cells.
Sun Z, Wang CY, Lawson DA, Kwek S, Velozo HG, Owyong M, Lai MD, Fong L, Wilson M, Su H, Werb Z, Cooke DL. Sun Z, et al. Oncotarget. 2017 Dec 29;9(13):10945-10961. doi: 10.18632/oncotarget.23760. eCollection 2018 Feb 16. Oncotarget. 2017. PMID: 29541388 Free PMC article.
References
- Hicks SC, Teng M, Irizarry RA. On the widespread and critical impact of systematic bias and batch effects in single-cell RNA-Seq data. bioRxiv. 2015. doi: http://dx.doi.org/10.1101/025528. - DOI
Publication types
MeSH terms
Grants and funding
- U54 AI117924/AI/NIAID NIH HHS/United States
- R01 GM102756/GM/NIGMS NIH HHS/United States
- R01GM102756/GM/NIGMS NIH HHS/United States
- T32 LM012413/LM/NLM NIH HHS/United States
- T32 HL083806/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources