Arf-like GTPase Arl8: Moving from the periphery to the center of lysosomal biology - PubMed (original) (raw)
Review
Arf-like GTPase Arl8: Moving from the periphery to the center of lysosomal biology
Divya Khatter et al. Cell Logist. 2015.
Abstract
Lysosomes are dynamic organelles that not only mediate degradation of cellular substrates but also play critical roles in processes such as cholesterol homeostasis, plasma membrane repair, antigen presentation, and cell migration. The small GTPase Arl8, a member of Arf-like (Arl) family of proteins, has recently emerged as a crucial regulator of lysosome positioning and membrane trafficking toward lysosomes. Through interaction with its effector SKIP, the human Arl8 paralog (Arl8b) mediates kinesin-1 dependent motility of lysosomes on microtubule tracks toward the cell periphery. Arl8b-mediated kinesin-driven motility is also implicated in regulating lytic granule polarization in NK cells, lysosome tubulation in macrophages, cell spreading, and migration. Moreover, Arl8b regulates membrane traffic toward lysosomes by recruiting subunits of the HOPS complex, a multi-subunit tethering complex that mediates endo-lysosome fusion. Here we provide a brief review on this recently characterized lysosomal GTPase and summarize the studies focusing on its known functions in regulating lysosomal motility and delivery of endocytic cargo to the lysosomes. We also explore the role of human Arl8b and its orthologs upon infection by intracellular pathogens.
Keywords: Arf-like GTPase; Arl8; BORC; HOPS; Motors; SKIP; autophagy; late endosome; lysosome; membrane trafficking.
Figures
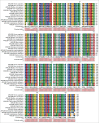
Figure 1.
Conservation of Arl8 protein across species. Arl8 is a primitive GTPase which is highly conserved from protozoans to metazoans as well in plants. Multiple sequence alignment was constructed using CLC sequence viewer software and NCBI accession numbers of the protein sequences used for sequence comparison were as follows: Homo sapiens (man) Arl8a, NP_620150.1; Homo sapiens (man) Arl8b, NP_060654.1; Pan troglodytes (chimpanzee), XP_001142369.1; Canis lupus familiaris (dog), XP_853013.1; Bos taurus (cattle), NP_001039536.1; Gallus gallus (chicken), XP_414440.3; Mus musculus (mouse), NP_080287.1; Rattus norvegicus (rat), NP_001019503.1; Xenopus tropicalis (frog), NP_001190182.1; Danio rerio (zebrafish), XP_002663861.1; Caenorhabditis elegans (worm), NP_502791.1; Drosophila melanogaster (fruit fly), NP_649769.1; Arabidopsis thaliana (small flowering plant), NP_568553.1; and Oryza sativa (rice plant), NP_001048027.1.
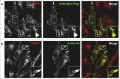
Figure 2.
Human Arl8 paralogs (Arl8a and Arl8b) localize to lysosomes. HeLa cells were transfected with either Arl8a-Myc-Flag (A) or Arl8b-HA (B) and analyzed for lysosomal localization by confocal microscopy. The figure indicates the peripheral recruitment of LAMP1-positive endosomes (red) in Arl8a/b transfected cells (green). Colocalized pixels are represented in the inset. Scale bar: 10 μm.
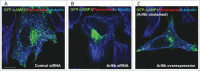
Figure 3.
Human Arl8b governs the distribution of lysosomes in mammalian cells. Control (A), Arl8b-silenced (B), and Arl8b-overexpressing (C) HeLa cells were transfected with GFP-LAMP1 and costained with antibodies to α-tubulin (blue) to label microtubules and pericentrin (red) to label MTOC. Knockdown of Arl8b results in lysosomal clustering at MTOC while its overexpression distributes lysosomes to the cell periphery. Scale bar: 10 μm.
Figure 4.
A model depicting the interplay of transport and fusion machinery operating at endosome-lysosome junction. Small GTPase Arl8b is recruited to lysosomal membranes by multi-subunit complex BORC. GTP-bound Arl8b interacts with its effectors: SKIP to mediate kinesin-dependent anterograde lysosomal movement and HOPS to accomplish vesicle (late endosome/phagosome) fusion at lysosomes.
Similar articles
- The small GTPase Arl8b regulates assembly of the mammalian HOPS complex on lysosomes.
Khatter D, Raina VB, Dwivedi D, Sindhwani A, Bahl S, Sharma M. Khatter D, et al. J Cell Sci. 2015 May 1;128(9):1746-61. doi: 10.1242/jcs.162651. Epub 2015 Apr 23. J Cell Sci. 2015. PMID: 25908847 Free PMC article. - ARL8 Relieves SKIP Autoinhibition to Enable Coupling of Lysosomes to Kinesin-1.
Keren-Kaplan T, Bonifacino JS. Keren-Kaplan T, et al. Curr Biol. 2021 Feb 8;31(3):540-554.e5. doi: 10.1016/j.cub.2020.10.071. Epub 2020 Nov 23. Curr Biol. 2021. PMID: 33232665 Free PMC article. - BORC Functions Upstream of Kinesins 1 and 3 to Coordinate Regional Movement of Lysosomes along Different Microtubule Tracks.
Guardia CM, Farías GG, Jia R, Pu J, Bonifacino JS. Guardia CM, et al. Cell Rep. 2016 Nov 15;17(8):1950-1961. doi: 10.1016/j.celrep.2016.10.062. Cell Rep. 2016. PMID: 27851960 Free PMC article. - Rab7: role of its protein interaction cascades in endo-lysosomal traffic.
Wang T, Ming Z, Xiaochun W, Hong W. Wang T, et al. Cell Signal. 2011 Mar;23(3):516-21. doi: 10.1016/j.cellsig.2010.09.012. Epub 2010 Sep 21. Cell Signal. 2011. PMID: 20851765 Review. - Mechanisms of lysosomal positioning and movement.
Cabukusta B, Neefjes J. Cabukusta B, et al. Traffic. 2018 Oct;19(10):761-769. doi: 10.1111/tra.12587. Epub 2018 Jul 17. Traffic. 2018. PMID: 29900632 Free PMC article. Review.
Cited by
- The Arf family GTPases: Regulation of vesicle biogenesis and beyond.
Li FL, Guan KL. Li FL, et al. Bioessays. 2023 Jun;45(6):e2200214. doi: 10.1002/bies.202200214. Epub 2023 Mar 30. Bioessays. 2023. PMID: 36998106 Free PMC article. Review. - Human CRB1 and CRB2 form homo- and heteromeric protein complexes in the retina.
Stehle IF, Imventarza JA, Woerz F, Hoffmann F, Boldt K, Beyer T, Quinn PM, Ueffing M. Stehle IF, et al. Life Sci Alliance. 2024 Apr 3;7(6):e202302440. doi: 10.26508/lsa.202302440. Print 2024 Jun. Life Sci Alliance. 2024. PMID: 38570189 Free PMC article. - Endosomal traffic disorders: a driving force behind neurodegenerative diseases.
Dong J, Tong W, Liu M, Liu M, Liu J, Jin X, Chen J, Jia H, Gao M, Wei M, Duan Y, Zhong X. Dong J, et al. Transl Neurodegener. 2024 Dec 24;13(1):66. doi: 10.1186/s40035-024-00460-7. Transl Neurodegener. 2024. PMID: 39716330 Free PMC article. Review. - Salmonella exploits the host endolysosomal tethering factor HOPS complex to promote its intravacuolar replication.
Sindhwani A, Arya SB, Kaur H, Jagga D, Tuli A, Sharma M. Sindhwani A, et al. PLoS Pathog. 2017 Oct 30;13(10):e1006700. doi: 10.1371/journal.ppat.1006700. eCollection 2017 Oct. PLoS Pathog. 2017. PMID: 29084291 Free PMC article. - Altered expression of vesicular trafficking machinery in prostate cancer affects lysosomal dynamics and provides insight into the underlying biology and disease progression.
Nturubika BD, Guardia CM, Gershlick DC, Logan JM, Martini C, Heatlie JK, Lazniewska J, Moore C, Lam GT, Li KL, Ung BS, Brooks RD, Hickey SM, Bert AG, Gregory PA, Butler LM, O'Leary JJ, Brooks DA, Johnson IRD. Nturubika BD, et al. Br J Cancer. 2024 Nov;131(8):1263-1278. doi: 10.1038/s41416-024-02829-x. Epub 2024 Aug 31. Br J Cancer. 2024. PMID: 39217195 Free PMC article.
References
- Luzio JP, Parkinson MD, Gray SR, Bright NA. The delivery of endocytosed cargo to lysosomes. Biochem Soc Trans 2009; 37:1019-21; PMID:19754443; http://dx.doi.org/ 10.1042/BST0371019 - DOI - PubMed
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069-75; PMID:18305538; http://dx.doi.org/ 10.1038/nature06639 - DOI - PMC - PubMed
- Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol 2012; 22:407-17; PMID:22748206; http://dx.doi.org/ 10.1016/j.tcb.2012.05.006 - DOI - PMC - PubMed
- Wartosch L, Bright NA, Luzio JP. Lysosomes. Curr Biol 2015; 25:R315-6; PMID:25898096; http://dx.doi.org/ 10.1016/j.cub.2015.02.027 - DOI - PubMed
- Futerman AH, van Meer G. The cell biology of lysosomal storage disorders. Nat Rev Mol Cell Biol 2004; 5:554-65; PMID:15232573; http://dx.doi.org/ 10.1038/nrm1423 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources