Rhizosphere Microbiomes of European + Seagrasses Are Selected by the Plant, But Are Not Species Specific - PubMed (original) (raw)
Rhizosphere Microbiomes of European + Seagrasses Are Selected by the Plant, But Are Not Species Specific
Catarina Cúcio et al. Front Microbiol. 2016.
Abstract
Seagrasses are marine flowering plants growing in soft-body sediments of intertidal and shallow sub-tidal zones. They play an important role in coastal ecosystems by stabilizing sediments, providing food and shelter for animals, and recycling nutrients. Like other plants, seagrasses live intimately with both beneficial and unfavorable microorganisms. Although much is known about the microbiomes of terrestrial plants, little is known about the microbiomes of seagrasses. Here we present the results of a detailed study on the rhizosphere microbiome of seagrass species across the North-eastern Atlantic Ocean: Zostera marina, Zostera noltii, and Cymodocea nodosa. High-resolution amplicon sequencing of 16S rRNA genes showed that the rhizobiomes were significantly different from the bacterial communities of surrounding bulk sediment and seawater. Although we found no significant differences between the rhizobiomes of different seagrass species within the same region, those of seagrasses in different geographical locations differed strongly. These results strongly suggest that the seagrass rhizobiomes are shaped by plant metabolism, but not coevolved with their host. The core rhizobiome of seagrasses includes mostly bacteria involved in the sulfur cycle, thereby highlighting the importance of sulfur-related processes in seagrass ecosystems.
Keywords: 16S rRNA; amplicon sequencing; marine bacteria; plant–microbe interactions; rhizosphere; seagrass microbiome; sulfur bacteria; sulfur cycle.
Figures
FIGURE 1
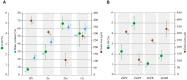
Abiotic characterization of sediments from the study areas in Portugal and France. Mean percentages of water content, loss on ignition (LOI), as well as median grain size are shown for bulk sediments (BS) and sediments of Z. noltii (Zn), Z. marina (Zm), and C. nodosa (Cn) sampled in Portugal (A). Comparison of LOI and grain size Zostera spp. sediments between Portugal and France. The data of ZnFr and ZmFr were obtained from Ouisse et al. (2010, 2012) (B). Bars indicate standard deviations, and letters reveal presence/absence of significant relationships between samples on each analysis with a significance level of 0.05, as determined by Tukey’s HSD post hoc test.
FIGURE 2
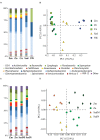
Comparison of bacterial communities of the seagrass rhizosphere (Z. marina, Zm; Z. noltii, ZN; C. nodosa, CN) and the surrounding environment (bulk sediment, Sed; and seawater (SW) from Portugal and France. Bulk sediments from the surroundings of Z. marina and Z. noltii are distinguished (SedM and SedN, respectively). Shown are the bacterial community compositions (average of five independent samples) from (A) Portugal and (C) France, and principal component analysis (PCA) of the bacterial communities from (B) Portugal and (D) France. Both PCAs show a clear separation between the bacterial communities of the rhizosphere and those from the surrounding environment. Only classes represented by an average abundance above 0.5 % are shown on the legend of the bar graphs (A,C), and the percentage of community variance explained by each axis is indicated in parentheses (B,D).
FIGURE 3
Box plots showing the relative abundance of sequences of the most abundant groups in the rhizobiome of seagrasses from Portugal and France, and the bacterial communities from the surrounding environment. Indicated are bulk sediments from Portugal (sedPT) and France (sedFR); seagrasses from Portugal (sgPT) and France (sgFR); seawater from Portugal (swPT). The top of the box indicates the third quartile, the bottom the first quartile, and the line in the middle is the median. The star indicates the mean of the data, crosses are outliers, and the whiskers represent error bars. (A) Gammaproteobacteria, (B) Deltaproteobacteria, (C) Epsilonproteobacteria, (D) Bacteroidia, (E) Alphaproteobacteria, and (F) Flavobacteriia.
FIGURE 4
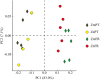
Principal component analysis (PCA) plot comparing bacterial communities of the rhizosphere of the seagrasses Z. marina (Zm) and Z. noltii (Zn), from Portugal (PT) and France (FR). Percentage of community variance explained by each axis is indicated in parentheses.
FIGURE 5
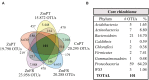
Venn diagram showing the core rhizobiome of all seagrasses studied. (A) Zostera marina (Zm-), Z. noltii (Zn-), both from Portugal (-PT) and France (-FR), and C. nodosa from Portugal (CnPT). The total number of OTUs clustered at a similarity level of 97% is represented under the label of each sample. The number of unique OTUs and OTUs shared between each combination of two samples is also shown. (B) Summarized taxonomic composition of the core rhizobiome, and respective number of OTUs present in each phylum. The relative number of the OTUs present in each phylum is also shown, indicated as percentage of sequences. A complete overview of the identity of the 101 OTUs that comprise the core rhizobiome of seagrasses is presented in Supplementary Table S3.
FIGURE 6
Box plots showing the relative abundance of sequences of the most abundant bacterial groups involved in sulfur processes. (A) Desulfobacteraceae, (B) Desulfobulbaceae, (C) Chromatiaceae, (D) Thiotrichaceae, (E) Campylobacteraceae, and (F) Helicobacteraceae. Indicated are bulk sediments from Portugal (sedPT) and France (sedFR); seagrasses from Portugal (sgPT) and France (sgFR); seawater from Portugal (swPT). The top of the box indicates the third quartile, the bottom the first quartile, and the line in the middle is the median. The star indicates the mean of the data, crosses are outliers, and the whiskers represent error bars.
Similar articles
- Rhizosphere microbiomes are closely linked to seagrass species: a comparative study of three coastal seagrasses.
Sun H, Liu X, Wang T, Liu S, Zhang R, Guo X, Yu Z, Zhao Y, Shen P, Zhang Y. Sun H, et al. Appl Environ Microbiol. 2024 Dec 18;90(12):e0175424. doi: 10.1128/aem.01754-24. Epub 2024 Nov 6. Appl Environ Microbiol. 2024. PMID: 39503478 Free PMC article. - Recovery and Community Succession of the Zostera marina Rhizobiome after Transplantation.
Wang L, English MK, Tomas F, Mueller RS. Wang L, et al. Appl Environ Microbiol. 2021 Jan 15;87(3):e02326-20. doi: 10.1128/AEM.02326-20. Print 2021 Jan 15. Appl Environ Microbiol. 2021. PMID: 33187993 Free PMC article. - Global Diversity and Biogeography of the Zostera marina Mycobiome.
Ettinger CL, Vann LE, Eisen JA. Ettinger CL, et al. Appl Environ Microbiol. 2021 May 26;87(12):e0279520. doi: 10.1128/AEM.02795-20. Epub 2021 May 26. Appl Environ Microbiol. 2021. PMID: 33837008 Free PMC article. - The Seagrass Holobiont and Its Microbiome.
Ugarelli K, Chakrabarti S, Laas P, Stingl U. Ugarelli K, et al. Microorganisms. 2017 Dec 15;5(4):81. doi: 10.3390/microorganisms5040081. Microorganisms. 2017. PMID: 29244764 Free PMC article. Review. - Antioxidant and Anti-Inflammatory Properties of Four Native Mediterranean Seagrasses: A Review of Bioactive Potential and Ecological Context.
Vasarri M, De Marchi L, Pretti C, Barletta E, Degl'Innocenti D. Vasarri M, et al. Mar Drugs. 2025 May 12;23(5):206. doi: 10.3390/md23050206. Mar Drugs. 2025. PMID: 40422796 Free PMC article. Review.
Cited by
- Seagrass-mediated rhizosphere redox gradients are linked with ammonium accumulation driven by diazotrophs.
Brodersen KE, Mosshammer M, Bittner MJ, Hallstrøm S, Santner J, Riemann L, Kühl M. Brodersen KE, et al. Microbiol Spectr. 2024 Apr 2;12(4):e0333523. doi: 10.1128/spectrum.03335-23. Epub 2024 Mar 1. Microbiol Spectr. 2024. PMID: 38426746 Free PMC article. - A Tight Interaction between the Native Seagrass Cymodocea nodosa and the Exotic Halophila stipulacea in the Aegean Sea Highlights Seagrass Holobiont Variations.
Conte C, Apostolaki ET, Vizzini S, Migliore L. Conte C, et al. Plants (Basel). 2023 Jan 11;12(2):350. doi: 10.3390/plants12020350. Plants (Basel). 2023. PMID: 36679063 Free PMC article. - Microbial communities on eelgrass (Zostera marina) thriving in Tokyo Bay and the possible source of leaf-attached microbes.
Iqbal MM, Nishimura M, Haider MN, Yoshizawa S. Iqbal MM, et al. Front Microbiol. 2023 Jan 6;13:1102013. doi: 10.3389/fmicb.2022.1102013. eCollection 2022. Front Microbiol. 2023. PMID: 36687565 Free PMC article. - Molecular Taxonomic Profiling of Bacterial Communities in a Gilthead Seabream (Sparus aurata) Hatchery.
Califano G, Castanho S, Soares F, Ribeiro L, Cox CJ, Mata L, Costa R. Califano G, et al. Front Microbiol. 2017 Feb 14;8:204. doi: 10.3389/fmicb.2017.00204. eCollection 2017. Front Microbiol. 2017. PMID: 28261166 Free PMC article. - Diversity of marine bacteria growing on leachates from virgin and weathered plastic: Insights into potential degraders.
Romera-Castillo C, Birnstiel S, Sebastián M. Romera-Castillo C, et al. Environ Microbiol Rep. 2024 Jun;16(3):e13305. doi: 10.1111/1758-2229.13305. Environ Microbiol Rep. 2024. PMID: 38923399 Free PMC article.
References
- Apostolopoulou M. V., Monteyne E., Krikonis K., Pavlopoulos K., Roose P., Dehairs F. (2014). Monitoring polycyclic aromatic hydrocarbons in the Northeast Aegean Sea using Posidonia oceanica seagrass and synthetic passive samplers. Mar. Pollut. Bull. 87 338–344. 10.1016/j.marpolbul.2014.07.051 - DOI - PubMed
- Asmus R. M., Sprung M., Asmus H. (2000). Nutrient fluxes in intertidal communities of a South European lagoon (Ria Formosa) – similarities and differences with a northern Wadden Sea bay (Sylt-Rømø Bay). Hydrobiologia 436 217–235. 10.1023/A:1026542621512 - DOI
- Bachelet G., Guillou J., Labourg P. J. (1992). “Adult-larval and juvenile interactions in the suspension-feeding bivalve, Cerastoderma edule (L.): field observations and experiments,” in Marine Eutrophication and Population Dynamics, eds Colombo G., Ferrari I., Ceccherelli V. U., Rossi R. (Fredensborg: Olsen & Olsen; ), 175–182.
LinkOut - more resources
Full Text Sources
Other Literature Sources