PD-L1 and PD-L2 Genetic Alterations Define Classical Hodgkin Lymphoma and Predict Outcome - PubMed (original) (raw)
. 2016 Aug 10;34(23):2690-7.
doi: 10.1200/JCO.2016.66.4482. Epub 2016 Apr 11.
Ranjana H Advani 1, Azra H Ligon 1, Yasodha Natkunam 1, Robert A Redd 1, Heather Homer 1, Courtney F Connelly 1, Heather H Sun 1, Sarah E Daadi 1, Gordon J Freeman 1, Philippe Armand 1, Bjoern Chapuy 1, Daphne de Jong 1, Richard T Hoppe 1, Donna S Neuberg 1, Scott J Rodig 1, Margaret A Shipp 2
Affiliations
- PMID: 27069084
- PMCID: PMC5019753
- DOI: 10.1200/JCO.2016.66.4482
PD-L1 and PD-L2 Genetic Alterations Define Classical Hodgkin Lymphoma and Predict Outcome
Margaretha G M Roemer et al. J Clin Oncol. 2016.
Abstract
Purpose: Classical Hodgkin lymphomas (cHLs) include small numbers of malignant Reed-Sternberg cells within an extensive but ineffective inflammatory/immune cell infiltrate. In cHL, chromosome 9p24.1/PD-L1/PD-L2 alterations increase the abundance of the PD-1 ligands, PD-L1 and PD-L2, and their further induction through Janus kinase 2-signal transducers and activators of transcription signaling. The unique composition of cHL limits its analysis with high-throughput genomic assays. Therefore, the precise incidence, nature, and prognostic significance of PD-L1/PD-L2 alterations in cHL remain undefined.
Methods: We used a fluorescent in situ hybridization assay to evaluate CD274/PD-L1 and PDCD1LG2/PD-L2 alterations in 108 biopsy specimens from patients with newly diagnosed cHL who were treated with the Stanford V regimen and had long-term follow-up. In each case, the frequency and magnitude of 9p24.1 alterations-polysomy, copy gain, and amplification-were determined, and the expression of PD-L1 and PD-L2 was evaluated by immunohistochemistry. We also assessed the association of 9p24.1 alterations with clinical parameters, which included stage (early stage I/II favorable risk, early stage unfavorable risk, advanced stage [AS] III/IV) and progression-free survival (PFS).
Results: Ninety-seven percent of all evaluated cHLs had concordant alterations of the PD-L1 and PD-L2 loci (polysomy, 5% [five of 108]; copy gain, 56% [61 of 108]; amplification, 36% [39 of 108]). There was an association between PD-L1 protein expression and relative genetic alterations in this series. PFS was significantly shorter for patients with 9p24.1 amplification, and the incidence of 9p24.1 amplification was increased in patients with AS cHL.
Conclusion: PD-L1/PD-L2 alterations are a defining feature of cHL. Amplification of 9p24.1 is more common in patients with AS disease and associated with shorter PFS in this series. Further analyses of 9p24.1 alterations in patients treated with standard cHL induction regimens or checkpoint blockade are warranted.
© 2016 by American Society of Clinical Oncology.
Conflict of interest statement
Authors' disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
Figures
Fig 1.
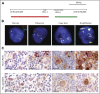
Genetic and immunohistochemical analyses of the PD-L1 and PD-L2 loci and PD-1 ligand expression. (A) Location and color labeling of the bacterial artificial chromosome (chr) clones on 9p24.1 used for fluorescent in situ hybridization (FISH). RP11-599H20 including PD-L1, labeled red. RP11-635N21 including PD-L2, labeled green. (B) Representative images of FISH results for the various categories. PD-L1 in red, PD-L2 in green, fused (F) signals in yellow, and centromeric probe (CEP9) in aqua (A). In these images, disomy reflects 2A:2F; polysomy, 3A:3F; copy gain, 3A:6F; and amplification, 15+F. (C) The top panel shows PD-L1 (brown)/PAX5 (red) immunohistochemistry (IHC) in the classical Hodgkin lymphoma (cHL) cases with 9p24.1 disomy, polysomy, copy gain, and amplification from (B). The bottom panel shows PD-L2 (brown)/pSTAT3 (red) IHC in the same cHL cases. Scale bar = 50 μm.
Fig 2.
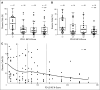
The spectrum of 9p24.1 alterations in classical Hodgkin lymphoma (cHL). (A) 9p24.1 alterations in evaluated cHLs. The cHLs are classified by the highest observed level of 9p24.1 alteration in Reed-Sternberg (RS) cells: polysomy, copy gain, or amplification (top). Individual tumors are depicted as columns on the _x_-axis. In each cHL, the percentage of RS cells with 9p24.1 disomy (black), polysomy (light pink), copy gain (pink), and/or amplification (red) is shown on the _y_-axis. (B) Percentage of RS cells with residual 9p24.1 disomy in cHLs classified by 9p24.1 alterations, as represented as box-and-whisker plots, showing minimum, first quartile, median, third quartile, and maximum. cHLs with 9p24.1 polysomy, copy gain, and amplification have significantly different percentages of residual RS cells with normal (disomic) 9p24.1 copy numbers. P < .001, Kruskal-Wallis test. (C) Association of PD-L1 protein expression and 9p24.1 copy number alterations. Residual 9p24.1 disomy is depicted on the _y_-axis; PD-L1 immunohistochemistry (IHC) H-score (in quartiles) is shown on the _x_-axis. Quartiles are indicated by dashed lines. A locally weighted polynomial regression line is shown in black. A highly significant decrease in percentage of residual 9p24.1 disomic cells in cHLs with a higher PD-L1 IHC H-score is shown. P = .005, Kruskal-Wallis test.
Fig 3.
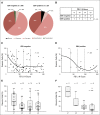
Clinical and genetic predictors of progression-free survival (PFS). (A) PFS by clinical stage in patients with classical Hodgkin lymphoma (cHL), early stage favorable (ES-F; n = 33), early stage unfavorable (ES-U; n = 41), and advanced stage (AS; n = 34). P = .002, log-rank test. (B) PFS by 9p24.1 alterations in patients with cHL (disomy, n = 1; polysomy, n = 5; copy gain, n = 61; amplification, n = 39; translocation, n = 2; P < .001, log-rank test). (C) Percentage of patients with 9p24.1 disomy (1%), polysomy (5%), copy gain (56%), amplification (36%), and translocation (1%) in the current series. (D) Frequency of 9p24.1 alterations (polysomy, copy gain, amplification, translocation, or disomy) by clinical stage (ES-F, ES-U, and AS) in this series. The incidence of 9p24.1 amplification is significantly different in clinically staged patients (ES-F, 24%; ES-U, 34%; AS, 50%; P = .024, Kruskal-Wallis test).
Fig A1.
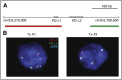
Chromosomal rearrangements in classical Hodgkin lymphoma (cHL). (A) Location and color labeling of the bacterial artificial chromosome (chr) clones on 9p24.1 used for fluorescent in situ hybridization (FISH) in translocation 2 (Tx #2). RP11-599H20 including PD-L1, labeled red; RP11-610G2 downstream of PD-L2, labeled green. For labeling of bacterial artificial chromosome clones used for translocation 1 (Tx #1), see Figure 1A. (B) FISH analyses of the cHL cases with chromosomal rearrangements. PD-L1 in red, PD-L2 in green, and centromeric probe (CEP9) in aqua. Arrows indicate the rearranged allele.
Fig A2.
Association of PD-L1/PD-L2 protein expression and 9p24.1 copy number alterations. (A) Percentage of 9p24.1 disomic cells in each of the PD-L1 immunohistochemistry (IHC) H-score quartiles. The _y_-axis shows the percentage of residual 9p24.1 disomic cells; the _x_-axis shows PD-L1 IHC H-score in quartiles. A statistically significant decrease in the percentage of normal (disomic) cells in the H-score quartiles was found. P = .005, Kruskal-Wallis test. (B) Percentage of residual 9p24.1 disomic cells in each of the PD-L2 IHC H-score quartiles. The _y_-axis shows the percentage of residual 9p24.1 disomic cells; the _x_-axis shows the PD-L2 IHC H-score in quartiles. The percentage of residual 9p24.1 disomic cells is statistically different in the quartiles. P = .006, Kruskal-Wallis test. (C) Percentage of residual 9p24.1 disomic cells (_y_-axis) and PD-L2 IHC H-score (_x_-axis) plotted for individual cases. Quartiles are indicated with dashed lines, and a trend line (locally weighted polynomial regression line) is shown in black. q, quartile.
Fig A3.
Distribution of genetic alterations in patients with Epstein-Barr virus (EBV) –negative and EBV-positive classical Hodgkin lymphoma (cHL). (A) The status of PD-L1 and _PD-L2_—disomy, polysomy, copy gain, amplification, and translocation—in EBV-negative (n = 88) and EBV-positive (n = 20) cHLs is visualized with a pie chart. (B) Distribution of EBV-negative and EBV-positive cases in the various PD-L1 immunohistochemistry (IHC) H-score quartiles. The proportion of EBV-positive cases increases as the H-score quartile category increases. P = .019, Kruskal-Wallis test. (C) and (D) Percentage of 9p24.1 residual disomic cells (_y_-axis) and PD-L1 IHC H-score (_x_-axis) plotted for individual EBV-negative and EBV-positive cHLs, respectively. Quartiles are indicated with dashed lines, and a trend line (locally weighted polynomial regression line) is shown in black. (E) and (F) Percentages of residual 9p24.1 disomic cells in EBV-negative and EBV-positive cases in the respective PD-L1 IHC H-score quartiles from (C) and (D) are plotted. The percentage of residual 9p24.1 disomic cells is significantly different in the respective H-score quartiles in EBV-negative cHLs in (E). P = .021, Kruskal-Wallis test. q, quartile.
Fig A4.
Progression-free survival (PFS) curves for the clinical risk groups with (gold) or without (blue) 9p24.1 amplification**:** early stage favorable (ES-F; n = 8 and n = 25, respectively), early stage unfavorable (ES-U; n = 14 and n = 27, respectively), and advanced stage (AS; n = 17 and n = 17, respectively).
Similar articles
- Classical Hodgkin Lymphoma with Reduced β2M/MHC Class I Expression Is Associated with Inferior Outcome Independent of 9p24.1 Status.
Roemer MG, Advani RH, Redd RA, Pinkus GS, Natkunam Y, Ligon AH, Connelly CF, Pak CJ, Carey CD, Daadi SE, Chapuy B, de Jong D, Hoppe RT, Neuberg DS, Shipp MA, Rodig SJ. Roemer MG, et al. Cancer Immunol Res. 2016 Nov;4(11):910-916. doi: 10.1158/2326-6066.CIR-16-0201. Epub 2016 Oct 13. Cancer Immunol Res. 2016. PMID: 27737878 Free PMC article. - PD-L1/PD-L2 genetic profile in the molecular cytogenetic classification of classic Hodgkin lymphoma.
García-Montenegro M, Narbaitz M, Metrebian MF, Pavlovsky A, Slavutsky I. García-Montenegro M, et al. Virchows Arch. 2025 May;486(5):1039-1047. doi: 10.1007/s00428-025-04047-z. Epub 2025 Feb 20. Virchows Arch. 2025. PMID: 39976683 - Expression pattern of PD-L1 and PD-L2 in classical Hodgkin lymphoma, primary mediastinal large B-cell lymphoma, and gray zone lymphoma.
Tanaka Y, Maeshima AM, Nomoto J, Makita S, Fukuhara S, Munakata W, Maruyama D, Tobinai K, Kobayashi Y. Tanaka Y, et al. Eur J Haematol. 2018 May;100(5):511-517. doi: 10.1111/ejh.13033. Epub 2018 Mar 1. Eur J Haematol. 2018. PMID: 29377256 - Signaling pathways and immune evasion mechanisms in classical Hodgkin lymphoma.
Liu WR, Shipp MA. Liu WR, et al. Hematology Am Soc Hematol Educ Program. 2017 Dec 8;2017(1):310-316. doi: 10.1182/asheducation-2017.1.310. Hematology Am Soc Hematol Educ Program. 2017. PMID: 29222272 Free PMC article. Review. - Signaling pathways and immune evasion mechanisms in classical Hodgkin lymphoma.
Liu WR, Shipp MA. Liu WR, et al. Blood. 2017 Nov 23;130(21):2265-2270. doi: 10.1182/blood-2017-06-781989. Blood. 2017. PMID: 29167175 Free PMC article. Review.
Cited by
- The Prognostic Value of Eight Immunohistochemical Markers Expressed in the Tumor Microenvironment and on Hodgkin Reed-Sternberg Cells in Pediatric Patients With Classical Hodgkin Lymphoma.
Zijtregtop EAM, Tromp I, Dandis R, Zwaan CM, Lam KH, Meyer-Wentrup FAG, Beishuizen A. Zijtregtop EAM, et al. Pathol Oncol Res. 2022 Aug 11;28:1610482. doi: 10.3389/pore.2022.1610482. eCollection 2022. Pathol Oncol Res. 2022. PMID: 36032657 Free PMC article. - The expression mechanism of programmed cell death 1 ligand 1 and its role in immunomodulatory ability of mesenchymal stem cells.
Chen Z, Yao MW, Ao X, Gong QJ, Yang Y, Liu JX, Lian QZ, Xu X, Zuo LJ. Chen Z, et al. Chin J Traumatol. 2024 Jan;27(1):1-10. doi: 10.1016/j.cjtee.2023.11.003. Epub 2023 Nov 23. Chin J Traumatol. 2024. PMID: 38065706 Free PMC article. Review. - Prognostic Markers within the Tumour Microenvironment in Classical Hodgkin Lymphoma.
Martynchyk A, Chowdhury R, Hawkes EA, Keane C. Martynchyk A, et al. Cancers (Basel). 2023 Oct 30;15(21):5217. doi: 10.3390/cancers15215217. Cancers (Basel). 2023. PMID: 37958391 Free PMC article. Review. - The potential of soluble programmed death-ligand 1 (sPD-L1) as a diagnosis marker for colorectal cancer.
Shao W, Xu Y, Lin S, Gao J, Gao J, Wang H. Shao W, et al. Front Oncol. 2022 Aug 16;12:988567. doi: 10.3389/fonc.2022.988567. eCollection 2022. Front Oncol. 2022. PMID: 36052227 Free PMC article. - Targeting immune checkpoints in hematological malignancies.
Salik B, Smyth MJ, Nakamura K. Salik B, et al. J Hematol Oncol. 2020 Aug 12;13(1):111. doi: 10.1186/s13045-020-00947-6. J Hematol Oncol. 2020. PMID: 32787882 Free PMC article. Review.
References
- Gordon LI, Hong F, Fisher RI, et al. Randomized phase III trial of ABVD versus Stanford V with or without radiation therapy in locally extensive and advanced-stage Hodgkin lymphoma: An intergroup study coordinated by the Eastern Cooperative Oncology Group (E2496). J Clin Oncol. 2013;31:684–691. - PMC - PubMed
- Hoskin PJ, Lowry L, Horwich A, et al. Randomized comparison of the Stanford V regimen and ABVD in the treatment of advanced Hodgkin’s Lymphoma: United Kingdom National Cancer Research Institute Lymphoma Group Study ISRCTN 64141244. J Clin Oncol. 2009;27:5390–5396. - PubMed
- Advani RH, Hong F, Fisher RI, et al. Randomized phase III trial comparing ABVD plus radiotherapy with the Stanford V regimen in patients with stages I or II locally extensive, bulky mediastinal Hodgkin lymphoma: A subset analysis of the North American Intergroup E2496 trial. J Clin Oncol. 2015;33:1936–1942. - PMC - PubMed
- Armitage JO. Early-stage Hodgkin’s lymphoma. N Engl J Med. 2010;363:653–662. - PubMed
- Kuruvilla J, Keating A, Crump M. How I treat relapsed and refractory Hodgkin lymphoma. Blood. 2011;117:4208–4217. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials