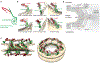Molecular architecture of the inner ring scaffold of the human nuclear pore complex - PubMed (original) (raw)
. 2016 Apr 15;352(6283):363-5.
doi: 10.1126/science.aaf0643.
Shyamal Mosalaganti 1, Alexander von Appen 1, Roman Teimer 2, Amanda L DiGuilio 3, William Wan 1, Khanh Huy Bui 4, Wim J H Hagen 1, John A G Briggs 5, Joseph S Glavy 3, Ed Hurt 2, Martin Beck 5
Affiliations
- PMID: 27081072
- PMCID: PMC8926079
- DOI: 10.1126/science.aaf0643
Molecular architecture of the inner ring scaffold of the human nuclear pore complex
Jan Kosinski et al. Science. 2016.
Abstract
Nuclear pore complexes (NPCs) are 110-megadalton assemblies that mediate nucleocytoplasmic transport. NPCs are built from multiple copies of ~30 different nucleoporins, and understanding how these nucleoporins assemble into the NPC scaffold imposes a formidable challenge. Recently, it has been shown how the Y complex, a prominent NPC module, forms the outer rings of the nuclear pore. However, the organization of the inner ring has remained unknown until now. We used molecular modeling combined with cross-linking mass spectrometry and cryo-electron tomography to obtain a composite structure of the inner ring. This architectural map explains the vast majority of the electron density of the scaffold. We conclude that despite obvious differences in morphology and composition, the higher-order structure of the inner and outer rings is unexpectedly similar.
Copyright © 2016, American Association for the Advancement of Science.
Figures
Fig. 1.. Composite structure of the human NPC.
(A) Overview of the composite structure of the entire NPC, in which previous structural assignments in the outer rings (9, 13) are represented together with the assignments in the IR that were undertaken in this study (details are shown in fig. S2). Unassigned density is shown in cyan; the nuclear ring is facing up. (B) Zoomed-in view of the IR region framed in (A). High-resolution structures (colored ribbons) are shown in the context of the tomographic map (transparent isosurface). (C) Conceptual outline of the NPC architecture. Inner (gray) and outer (orange) copies of the Ycomplex (top and bottom) and the IR core module (middle) are shown in comparison.The configurations of the outer and inner copies of the IR core module are shown enlarged on the right. FG repeat domains can readily reach out into the central channel (F, phenylalanine; G, glycine).
Fig. 2.. Cross-linking mass spectrometry confirms intraand inter-subcomplex interactions of the IR architecture.
(A) Primary schematic of the IR proteins, showing the cross-links obtained by mass spectrometry. Inter-protein cross-links are shown in black, intra-protein links in light purple, and homodimeric cross-links (linking the same residues of a protein) in red. For clarity, Nup53 is not drawn to scale. Colored areas within the primary structures mark domains and motifs as individually labeled (residues are indicated below). A larger version is shown in fig. S6, and table S1 provides details on the cross-link data sets. The cross-links [blue lines in (B) to (D)] confirm specific features suggested by the composite structure, such as (B) head-to-tail arrangement of the inner and outer copy of Nup93, (C) interaction between Nup93 and Nup205, and (D) positioning of the Nup54 ferredoxin-like domain within the Nup62 complex.
Fig. 3.. Simple and highly similar organizational principles govern the architecture of the inner and outer rings of the NPC.
(A) Schematic representation of the branching motif formed by the different heterodimers. (B) Structural similarity of the Nup155-Nup93 dimer to the Nup107-Nup133 and Nup120-Nup85 dimers. The structures are shown in low-resolution representation within the corresponding segments of the tomographic map. Homologous Nups are colored identically. (C) Arrangement of the branching motifs within the NPC membrane-binding coat [homologous Nups are colored as in (B)]. (D) Schematic of the human NPC architecture (details are shown in figs. S9 to S11). Trailing lines indicate FG repeats.
Comment in
- Boundary Issues.
[No authors listed] [No authors listed] Cell. 2016 May 5;165(4):759-61. doi: 10.1016/j.cell.2016.04.054. Cell. 2016. PMID: 27153485 No abstract available.
Similar articles
- In situ structural analysis of the human nuclear pore complex.
von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, Buczak K, Mosalaganti S, Hagen W, Andres-Pons A, Lemke EA, Bork P, Antonin W, Glavy JS, Bui KH, Beck M. von Appen A, et al. Nature. 2015 Oct 1;526(7571):140-143. doi: 10.1038/nature15381. Epub 2015 Sep 23. Nature. 2015. PMID: 26416747 Free PMC article. - Architecture of the linker-scaffold in the nuclear pore.
Petrovic S, Samanta D, Perriches T, Bley CJ, Thierbach K, Brown B, Nie S, Mobbs GW, Stevens TA, Liu X, Tomaleri GP, Schaus L, Hoelz A. Petrovic S, et al. Science. 2022 Jun 10;376(6598):eabm9798. doi: 10.1126/science.abm9798. Epub 2022 Jun 10. Science. 2022. PMID: 35679425 Free PMC article. - Architecture of the symmetric core of the nuclear pore.
Lin DH, Stuwe T, Schilbach S, Rundlet EJ, Perriches T, Mobbs G, Fan Y, Thierbach K, Huber FM, Collins LN, Davenport AM, Jeon YE, Hoelz A. Lin DH, et al. Science. 2016 Apr 15;352(6283):aaf1015. doi: 10.1126/science.aaf1015. Epub 2016 Apr 14. Science. 2016. PMID: 27081075 Free PMC article. - Structure and Assembly of the Nuclear Pore Complex.
Hampoelz B, Andres-Pons A, Kastritis P, Beck M. Hampoelz B, et al. Annu Rev Biophys. 2019 May 6;48:515-536. doi: 10.1146/annurev-biophys-052118-115308. Epub 2019 Apr 3. Annu Rev Biophys. 2019. PMID: 30943044 Review. - The Structure of the Nuclear Pore Complex (An Update).
Lin DH, Hoelz A. Lin DH, et al. Annu Rev Biochem. 2019 Jun 20;88:725-783. doi: 10.1146/annurev-biochem-062917-011901. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883195 Free PMC article. Review.
Cited by
- TissUExM enables quantitative ultrastructural analysis in whole vertebrate embryos by expansion microscopy.
Steib E, Tetley R, Laine RF, Norris DP, Mao Y, Vermot J. Steib E, et al. Cell Rep Methods. 2022 Sep 30;2(10):100311. doi: 10.1016/j.crmeth.2022.100311. eCollection 2022 Oct 24. Cell Rep Methods. 2022. PMID: 36313808 Free PMC article. - Evolutionary trajectory for nuclear functions of ciliary transport complex proteins.
Ewerling A, May-Simera HL. Ewerling A, et al. Microbiol Mol Biol Rev. 2024 Sep 26;88(3):e0000624. doi: 10.1128/mmbr.00006-24. Epub 2024 Jul 12. Microbiol Mol Biol Rev. 2024. PMID: 38995044 Review. - A Quantitative Model for BicD2/Cargo Interactions.
Noell CR, Loftus KM, Cui H, Grewer C, Kizer M, Debler EW, Solmaz SR. Noell CR, et al. Biochemistry. 2018 Nov 20;57(46):6538-6550. doi: 10.1021/acs.biochem.8b00987. Epub 2018 Nov 5. Biochemistry. 2018. PMID: 30345745 Free PMC article. - Atomic force microscopy reveals structural variability amongst nuclear pore complexes.
Stanley GJ, Fassati A, Hoogenboom BW. Stanley GJ, et al. Life Sci Alliance. 2018 Aug 20;1(4):e201800142. doi: 10.26508/lsa.201800142. eCollection 2018 Aug. Life Sci Alliance. 2018. PMID: 30456374 Free PMC article. - Role of Computational Methods in Going beyond X-ray Crystallography to Explore Protein Structure and Dynamics.
Srivastava A, Nagai T, Srivastava A, Miyashita O, Tama F. Srivastava A, et al. Int J Mol Sci. 2018 Oct 30;19(11):3401. doi: 10.3390/ijms19113401. Int J Mol Sci. 2018. PMID: 30380757 Free PMC article. Review.
References
- Raices M, D’Angelo MA, Nat. Rev. Mol. Cell Biol 13, 687–699 (2012). - PubMed
- Hoelz A, Debler EW, Blobel G, Annu. Rev. Biochem 80, 613–643 (2011). - PubMed
- Hurt E, Beck M, Curr. Opin. Cell Biol 34, 31–38 (2015). - PubMed
- Vollmer B, Antonin W, Biol. Chem 395, 515–528 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources