Architecture of the symmetric core of the nuclear pore - PubMed (original) (raw)
. 2016 Apr 15;352(6283):aaf1015.
doi: 10.1126/science.aaf1015. Epub 2016 Apr 14.
Tobias Stuwe 1, Sandra Schilbach 1, Emily J Rundlet 1, Thibaud Perriches 1, George Mobbs 1, Yanbin Fan 1, Karsten Thierbach 1, Ferdinand M Huber 1, Leslie N Collins 1, Andrew M Davenport 1, Young E Jeon 1, André Hoelz 2
Affiliations
- PMID: 27081075
- PMCID: PMC5207208
- DOI: 10.1126/science.aaf1015
Architecture of the symmetric core of the nuclear pore
Daniel H Lin et al. Science. 2016.
Abstract
The nuclear pore complex (NPC) controls the transport of macromolecules between the nucleus and cytoplasm, but its molecular architecture has thus far remained poorly defined. We biochemically reconstituted NPC core protomers and elucidated the underlying protein-protein interaction network. Flexible linker sequences, rather than interactions between the structured core scaffold nucleoporins, mediate the assembly of the inner ring complex and its attachment to the NPC coat. X-ray crystallographic analysis of these scaffold nucleoporins revealed the molecular details of their interactions with the flexible linker sequences and enabled construction of full-length atomic structures. By docking these structures into the cryoelectron tomographic reconstruction of the intact human NPC and validating their placement with our nucleoporin interactome, we built a composite structure of the NPC symmetric core that contains ~320,000 residues and accounts for ~56 megadaltons of the NPC's structured mass. Our approach provides a paradigm for the structure determination of similarly complex macromolecular assemblies.
Copyright © 2016, American Association for the Advancement of Science.
Conflict of interest statement
The authors declare no financial conflicts of interest.
Figures
Fig. 1. Reconstitution of NPC symmetric core protomers
(A) Cross-sectional conceptual schematic of the NPC. CNCs, colored yellow, form outer rings above the membrane on the nuclear and cytoplasmic faces. Adaptor and channel nucleoporins, colored orange and red, respectively, form concentric cylinders in the inner ring. Asymmetric nucleoporins decorate the cytoplasmic and nuclear faces of the NPC. (B) Domain organization of nucleoporins used in biochemical reconstitution experiments. Black lines indicate the construct boundaries used. Domains are drawn as boxes colored according to the legend at the bottom right for observed or predicted folds. Cartoons to the right are used throughout the text to represent the nucleoporins. Some nucleoporins form stable complexes (CNC, coat nucleoporin complex; CFC cytoplasmic filament nucleoporin complex; CNT, channel nucleoporin hetero-trimer) and are drawn as a unit. (C) Cartoon of experimental setup in panels (D–G). The CNC was preincubated with IRC/Nup188 complex in the presence or absence of Nup145N. Complex formation was only observed in the presence of Nup145N. (D, E) Reconstitution of NPC symmetric core protomers containing the CNC-hexamer and IRC is dependent on Nup145N. Identical experiments were performed in the (D) presence or (E) absence of Nup145N. Size-exclusion chromatography coupled to multiangle light scattering (SEC-MALS) profiles of the complexes in isolation (red and blue) and after preincubation (green) are shown. Representative Coomassie-stained SDS-PAGE gel slices for the peak fractions are shown with a colored arrow above the chromatogram indicating the resolved peak fraction. Measured molecular masses are indicated for each peak. Cartoons below the gel slices illustrate the respective complexes. (F, G) Reconstitution of NPC symmetric core protomers containing the CNC-hexamer and Nup188-complex is dependent on Nup145N. Identical experiments were performed in the (F) presence or (G) absence of Nup145N. Complete SDS-PAGE gels for all panels are shown in fig. S4.
Fig. 2. Biochemical analysis of the interactions mediating NPC symmetric core assembly
(A) Scaffold domains of Nup192, Nup188, Nup170, and Nic96SOL do not interact with each other. SEC-MALS profiles of the individual scaffolds alone (blue, purple, orange, and green) and after their preincubation with each other (black) are shown. Measured molecular masses are indicated for each peak. (B, C) Linker nucleoporins Nup53 and Nup145N mediate scaffold nucleoporin assembly. SEC-MALS profiles of a scaffold nucleoporin mixture (blue or purple), a linker nucleoporin mixture (red), and after their preincubation with each other (black) are shown. Mixtures of scaffold nucleoporins contained either (B) Nup192 or (C) Nup188. Complete SDS-PAGE gels for all chromatograms are shown in fig. S7. (D, E) Interaction network between scaffold nucleoporins and (D) Nup53 or (E) Nup145N. Scaffold nucleoporins were tested in SEC-MALS interaction experiments for their ability to form hetero-dimeric complexes with Nup53 or Nup145N and their compatibility to also form hetero-trimeric complexes. Check marks indicate complexes that can form in SEC-MALS experiments, crosses indicate complexes that do not form, and dashes indicate complexes that were not tested. Complete SEC-MALS chromatograms and SDS-PAGE gels are shown in figs. S8 to S11. (F) Biochemical interaction map revealed by SEC-MALS interaction experiments. Minimal regions of Nup145N and Nup53 sufficient for binding to components of the NPC are depicted using colored bars and dashed lines between interacting regions. Interactions that map to the same regions on Nup145N and Nup53 do not occur simultaneously. Complete SEC-MALS chromatograms and SDS-PAGE gels are shown in figs S13 to S20. The nucleoporin schematics are according to Fig. 1B.
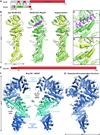
Fig. 3. Structural and biochemical analyses of the Nup170 interaction network
(A) Domain structures of Nup170 and Nup53. Black lines indicate fragments used for crystallization. (B) Crystal structure of the Nup170NTD•Nup53R3 complex shown in cartoon representation. A 90° rotation is shown on the right. The potential WF and ALPS membrane interaction motifs are indicated. (C) Close-up view of the interaction between Nup170 and Nup53. Nup170 and Nup53 residues involved in the interaction are labeled in orange and purple, respectively. (D) Graphic summary of mutational analysis of the Nup170NTD-Nup53R3 interaction. Nup170 is shown in surface representation from the same view as in panel (C). Residues are colored in red, orange, yellow, and green to indicate mutations that had a strong, moderate, weak, or no effect on binding, respectively. (E) Tabular summary of tested Nup170 mutants and their effect on Nup53 binding; (+++) wild-type binding, (++) moderately weakened binding, (+) weak binding, and (-) no binding. Mutations in Nup53 that were tested for binding are indicated by dots above the sequence using the same color code as in panel (D). Chromatograms and representative SDS-PAGE gels are shown in fig. S22. (F) Domain structures of Nup170 and Nup145N. Black lines indicate fragments used for crystallization. (G) Crystal structure of the Nup170CTD•Nup145NR3 complex shown in cartoon representation. (H) Close-up view of the interaction between Nup170 and Nup145N. Nup170 and Nup145N residues involved in the interaction are labeled in orange and cyan, respectively. (I) Graphic summary of mutational analysis of the Nup170CTD-Nup145NR3 interaction. Nup170 is shown in surface representation from the same view as in panel (H). Coloring is according to panel (D). (J) Summary of mutational analysis of the Nup170CTD-Nup145NR3 interaction, coloring and key are same as in panel (E). Chromatograms and representative SDS-PAGE gels are shown in fig. S23.
Fig. 4. Structural analysis of the inner ring complex nucleoporins Nic96 and Nup192
(A) Domain structures of Nic96 and Nup53. Black lines indicate fragments used for crystallization. (B) Crystal structures of apo Nic96SOL (yellow) and the Nic96SOL•Nup53R2 complex (green and purple) and their superposition are shown in cartoon representation. See also Movie 3. (C) Close-up view of the Nup53R2-binding site in Nic96SOL. For clarity, Nup53 is shown in ribbon representation. Nic96 and Nup53 residues involved in the interaction are labeled in green and purple, respectively. (D) Close-up view of the superposition of apo and Nup53-bound structures of Nic96SOL reveals minimal conformational changes. (E) Domain structure of Nup192. A black line indicates the fragment used for crystallization. (F) Crystal structure of Nup192ΔHEAD shown in cartoon representation. A 180° rotated view is shown on the right. Regions of Nup192 that were resolved in previous crystal structures are colored in shades of blue, while the region of the protein that was not included in previous crystallographic analyses is shaded cyan. (G) Structure of full-length Nup192 generated by superposing fragment crystal structures. See also fig. S31 and Movie 4.
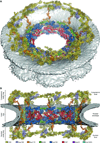
Fig. 5. Architecture of the NPC symmetric core
Composite structure of the NPC symmetric core generated by docking nucleoporin and nucleoporin complex crystal structures into the cryoET reconstruction of the intact human NPC (EMD-3103). The density corresponding to the nuclear envelope is shown as a gray surface. Proteins are color-coded according to the legend at the bottom. (A) View from above the cytoplasmic face and (B) a cross-sectional view from within the transport channel. Details are discussed in the text. See also figs. S34 to S37; S40; Movie 5.
Fig. 6. Architecture of the NPC spoke
(A) A single NPC symmetric core spoke is shown in cartoon representation from the same cross-sectional view as in Fig. 5B. Different shades of colors are used to indicate biochemically distinct dockings of the same protein. See also Movie 6. (B) Same view as in panel (A), but with Nup170 and the nucleoporins from the outer ring removed to highlight the organization of the four inner ring complexes (IRCs). (C) The membrane coat of the NPC. Nup192 and Nic96 molecules and CNTs have been removed for clarity. Contacts between the nuclear envelope and the ALPS motifs in Nup120, Nup133, and Nup170 are indicated by dots. (D) Schematic of the NPC spoke. The proteins corresponding to the nuclear side of the spoke are colored in gray, demonstrating the two-fold rotational symmetry relating the cytoplasmic and nuclear halves of each spoke. (E) Equatorial and peripheral IRCs adopt similar conformations. Identical views of the cytoplasmic peripheral IRC, the cytoplasmic equatorial IRCs, and their superposition are shown. For clarity, the equatorial IRC is colored in gray in the superposition. (F) Organization of FG repeats in the inner ring. Colored spheres indicate the positions of the N-termini of the three channel nucleoporins for all 32 CNT copies in the inner ring. Despite four distinct CNT positions in each spoke, the N-termini for the cytoplasmic and nuclear complexes are arranged in approximately the same plane. Thus, while not possessing true 16-fold symmetry, the CNT N-termini approximately form two 16-membered rings, which are indicated by solid and dashed lines. Notably, the FG repeats of the 16-membered CNT rings project circumferentially into the central transport channel with opposite directionality; cytoplasmic CNT ring (counterclockwise), nuclear CNT ring (clockwise). The CNT molecules for a single spoke are shown in cartoon representation to indicate the directionality of the FG repeats.
Fig. 7. Cylindrical organization of the NPC
(A) A top view of the composite structure of the NPC symmetric core viewed from the cytoplasm is shown (left). Coloring is according to Fig. 5. Schematic representation on the right illustrates the distinct concentric cylinders observed in the symmetric core. Cartoons of the linker nucleoporins and the unstructured NTE of Nic96 are shown to indicate how they span across multiple cylinders. Arrows indicate gaps between the inner ring spokes, which represent proposed paths for inner nuclear membrane protein transport. (B) Only a single contact is observed between the two adjacent inner ring spokes. A close-up view of the inter-spoke interface in the inner ring is shown in an orientation similar to Fig. 5A. Two adjacent spokes are depicted as gray and white surfaces, on the left and right, respectively. An equatorial Nup192 molecule (spoke 1) and a peripheral Nic96 molecule bound to Nup53 (spoke 2) are shown in cartoon representation. The Nup53 binding site on Nup192 is labeled and the proposed path of the peptide is drawn as a purple dashed line. (C) A schematic representation illustrating the concentric cylinder organization of the symmetric core shown from the side. The order and arrangement of the binding sites for the linker nucleoporins observed in the composite structure are depicted. The proposed inter-spoke interaction between Nup53 and Nup192 is drawn at the bottom.
Comment in
- Boundary Issues.
[No authors listed] [No authors listed] Cell. 2016 May 5;165(4):759-61. doi: 10.1016/j.cell.2016.04.054. Cell. 2016. PMID: 27153485 No abstract available.
Similar articles
- Architecture of the linker-scaffold in the nuclear pore.
Petrovic S, Samanta D, Perriches T, Bley CJ, Thierbach K, Brown B, Nie S, Mobbs GW, Stevens TA, Liu X, Tomaleri GP, Schaus L, Hoelz A. Petrovic S, et al. Science. 2022 Jun 10;376(6598):eabm9798. doi: 10.1126/science.abm9798. Epub 2022 Jun 10. Science. 2022. PMID: 35679425 Free PMC article. - Architecture of the cytoplasmic face of the nuclear pore.
Bley CJ, Nie S, Mobbs GW, Petrovic S, Gres AT, Liu X, Mukherjee S, Harvey S, Huber FM, Lin DH, Brown B, Tang AW, Rundlet EJ, Correia AR, Chen S, Regmi SG, Stevens TA, Jette CA, Dasso M, Patke A, Palazzo AF, Kossiakoff AA, Hoelz A. Bley CJ, et al. Science. 2022 Jun 10;376(6598):eabm9129. doi: 10.1126/science.abm9129. Epub 2022 Jun 10. Science. 2022. PMID: 35679405 Free PMC article. - Molecular architecture of the inner ring scaffold of the human nuclear pore complex.
Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJ, Briggs JA, Glavy JS, Hurt E, Beck M. Kosinski J, et al. Science. 2016 Apr 15;352(6283):363-5. doi: 10.1126/science.aaf0643. Science. 2016. PMID: 27081072 Free PMC article. - The Structure of the Nuclear Pore Complex (An Update).
Lin DH, Hoelz A. Lin DH, et al. Annu Rev Biochem. 2019 Jun 20;88:725-783. doi: 10.1146/annurev-biochem-062917-011901. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883195 Free PMC article. Review. - Functional architecture of the nuclear pore complex.
Grossman E, Medalia O, Zwerger M. Grossman E, et al. Annu Rev Biophys. 2012;41:557-84. doi: 10.1146/annurev-biophys-050511-102328. Annu Rev Biophys. 2012. PMID: 22577827 Review.
Cited by
- Allosteric modulation of nucleoporin assemblies by intrinsically disordered regions.
Blus BJ, Koh J, Krolak A, Seo HS, Coutavas E, Blobel G. Blus BJ, et al. Sci Adv. 2019 Nov 27;5(11):eaax1836. doi: 10.1126/sciadv.aax1836. eCollection 2019 Nov. Sci Adv. 2019. PMID: 31807700 Free PMC article. - Mapping the native organization of the yeast nuclear pore complex using nuclear radial intensity measurements.
Vallotton P, Rajoo S, Wojtynek M, Onischenko E, Kralt A, Derrer CP, Weis K. Vallotton P, et al. Proc Natl Acad Sci U S A. 2019 Jul 16;116(29):14606-14613. doi: 10.1073/pnas.1903764116. Epub 2019 Jul 1. Proc Natl Acad Sci U S A. 2019. PMID: 31262825 Free PMC article. - Biomimetic Nanomembranes: An Overview.
Jakšić Z, Jakšić O. Jakšić Z, et al. Biomimetics (Basel). 2020 May 29;5(2):24. doi: 10.3390/biomimetics5020024. Biomimetics (Basel). 2020. PMID: 32485897 Free PMC article. Review. - Control of yeast retrotransposons mediated through nucleoporin evolution.
Rowley PA, Patterson K, Sandmeyer SB, Sawyer SL. Rowley PA, et al. PLoS Genet. 2018 Apr 25;14(4):e1007325. doi: 10.1371/journal.pgen.1007325. eCollection 2018 Apr. PLoS Genet. 2018. PMID: 29694349 Free PMC article. - Sample Preparation for Electron Cryo-Microscopy of Macromolecular Machines.
Deniaud A, Kabasakal BV, Bufton JC, Schaffitzel C. Deniaud A, et al. Adv Exp Med Biol. 2024;3234:173-190. doi: 10.1007/978-3-031-52193-5_12. Adv Exp Med Biol. 2024. PMID: 38507207
References
- Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu. Rev. Biochem. 2011;80:613–643. - PubMed
- Kohler A, Hurt E. Gene regulation by nucleoporins and links to cancer. Mol. Cell. 2010;38:6–15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM007616/GM/NIGMS NIH HHS/United States
- ACB-12002/PHS HHS/United States
- AGM-12006/PHS HHS/United States
- R01-GM111461/GM/NIGMS NIH HHS/United States
- 5 T32 GM07616/GM/NIGMS NIH HHS/United States
- R01 GM117360/GM/NIGMS NIH HHS/United States
- R01 GM111461/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources