Slide-and-exchange mechanism for rapid and selective transport through the nuclear pore complex - PubMed (original) (raw)
Slide-and-exchange mechanism for rapid and selective transport through the nuclear pore complex
Barak Raveh et al. Proc Natl Acad Sci U S A. 2016.
Abstract
Nucleocytoplasmic transport is mediated by the interaction of transport factors (TFs) with disordered phenylalanine-glycine (FG) repeats that fill the central channel of the nuclear pore complex (NPC). However, the mechanism by which TFs rapidly diffuse through multiple FG repeats without compromising NPC selectivity is not yet fully understood. In this study, we build on our recent NMR investigations showing that FG repeats are highly dynamic, flexible, and rapidly exchanging among TF interaction sites. We use unbiased long timescale all-atom simulations on the Anton supercomputer, combined with extensive enhanced sampling simulations and NMR experiments, to characterize the thermodynamic and kinetic properties of FG repeats and their interaction with a model transport factor. Both the simulations and experimental data indicate that FG repeats are highly dynamic random coils, lack intrachain interactions, and exhibit significant entropically driven resistance to spatial confinement. We show that the FG motifs reversibly slide in and out of multiple TF interaction sites, transitioning rapidly between a strongly interacting state and a weakly interacting state, rather than undergoing a much slower transition between strongly interacting and completely noninteracting (unbound) states. In the weakly interacting state, FG motifs can be more easily displaced by other competing FG motifs, providing a simple mechanism for rapid exchange of TF/FG motif contacts during transport. This slide-and-exchange mechanism highlights the direct role of the disorder within FG repeats in nucleocytoplasmic transport, and resolves the apparent conflict between the selectivity and speed of transport.
Keywords: NMR; molecular dynamics; nuclear pore; nucleoporins; transport factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
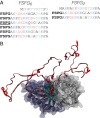
Fig. 1.
Simulated system. (A) Sequences of FG repeat model constructs. Bold, FSFG motif; underlined, motif bound at crystallographic binding site; blue, positive; red, negative; green, polar. (B) Starting conformation for bound FSFG6:NTF2 simulations, with the third FSFG motif initially bound at the crystallographic binding site (cyan space-fill), adapted from pdb-id 1GYB (66).
Fig. 2.
Biopolymer properties of FSFG repeats. (A) The secondary structures assigned to FSFG2 by the program STRIDE (88) throughout the 30-μs-long TIP4P-Ew simulation of FSFG2:NTF2. Helical or sheet secondary structures did not form during the simulations. See SI Appendix, Fig. S1 for secondary structure assignment in additional simulations. (B) Mean radius of gyration as a function of segment length in TIP4P-D or TIP4P-Ew simulations of FSFG6:NTF2 and FSFG2:NTF2 in the different water models. The Flory exponent was extracted by fitting to power law _n_F, where n is the segment length. (C) Distribution of distances between adjacent FG motifs in the presence of NTF2 using the TIP4P-D and TIP4P-Ew water models, computed in 1-Å bins. A bimodal Gaussian fit to the average over each water model is shown in space-fill for TIP4P-D (pink) and TIP4P-Ew (blue). (D) Change in end-to-end distance between consecutive FG motifs in TIP4P-D simulations, with fast fluctuations around the dominant peak on the nanosecond timescale.
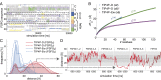
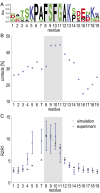
Fig. 3.
Binding dynamics to NTF2. (A) Sequence profile of the six FG repeats in FSFG6. (B) The fraction of simulation time in which different FSFG6 residues are in contact (<5 Å distance between any pair of heavy atoms) with NTF2 during the TIP4P-D simulation, averaged over all residues in corresponding repeat positions. (C) _R_2/_R_1 relaxation values measured experimentally by NMR (black) and in the FSFG6:NTF2 simulations (red). Sequence position 14 represents S alone. See SI Appendix, Fig. S6 for nonaveraged data for the entire sequence of FSFG.
Fig. 4.
Sliding of the FG motif residues along the crystallographic binding groove. (A) Superposition of the centers of mass of the FG motif residues (FSFG) at the crystallographic binding site from 10 independent TAMD simulations with the TIP4P-Ew water model (red spheres), and their principal components scaled by their eigenvalues (red arrows), showing a favorable lateral sliding mode of motion (PCA1). The NTF2 subunits are shown in blue and gray ribbon/surface representation. The trace of the center of mass of the same FG motif during the first 400 ns of the unbiased bound FSFG6:NTF2 simulation with the TIP4P-D water model is shown in a color gradient of spheres from its initial conformation (t = 0 ns; orange) to the final unbinding event (t = 400 ns; blue), as it slides along PCA1 (t = 200 ns; green) (Movies S3 and S4). (B) The principal component values during the unbinding in the unbiased FSFG6:NTF2 simulation shown in A and in Movies S3 and S4. (C) Occurrence of motions over 5 Å along the different principal components for RAMD simulations with decreasing random force magnitude (x axis). A random force of magnitude 0 kcal mol−1 Å−2 is equivalent to an unbiased simulation. Trend lines from a polynomial fit are plotted in gray.
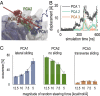
Fig. 5.
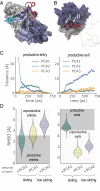
A sliding-and-exchange mechanism. (A) A strongly interacting motif slides out (motif II; pink to red) to a weakly interacting state as a competing motif (motif I; cyan to blue) slides in Movie S7 in the unbiased TIP4P-Ew simulation of FSFG2:NTF2. (B) Two individual FG motifs (magenta and red space-fill, indicated as I and II) accommodated simultaneously at the crystallographic binding site on the NTF2 dimer along the horizontal binding groove in the unbiased TIP4P-D simulation of FSFG6:NTF2. (C) A representative exchange trajectory (similar to other productive exchange trajectories which comprise 97% of all trajectories), showing the distance from the interaction site of either the displacing (Left) or displaced FG motif (Right), projected over each of the three principal components (as in Fig. 4_A_; SI Appendix, Table S2). See also Movie S9. (D) Distribution of productive vs. nonproductive exchange events in the targeted MD simulations (63), in which the displacing FG motif was targeted toward the interaction site from either the +PCA1, −PCA1, or +PCA2 directions. A productive exchange event requires productive entry (Left) and exit (Right), assessed here according to the final root-mean-square-deviation (RMSD) from the interaction site of either the displacing or displaced FG motifs, respectively.
Fig. 6.
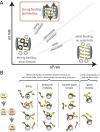
Balancing transport rate and selectivity. (A) Efficient nucleocytoplasmic transport requires strong binding affinity and a high off rate to guarantee fast exchange of TFs interacting with FG repeats. (B) Mechanisms for fast partial unbinding and rebinding in a dimensionally reduced energy landscape, compared with complete unbinding and rebinding.
Similar articles
- Intramolecular cohesion of coils mediated by phenylalanine--glycine motifs in the natively unfolded domain of a nucleoporin.
Krishnan VV, Lau EY, Yamada J, Denning DP, Patel SS, Colvin ME, Rexach MF. Krishnan VV, et al. PLoS Comput Biol. 2008 Aug 8;4(8):e1000145. doi: 10.1371/journal.pcbi.1000145. PLoS Comput Biol. 2008. PMID: 18688269 Free PMC article. - Nucleoporin's Like Charge Regions Are Major Regulators of FG Coverage and Dynamics Inside the Nuclear Pore Complex.
Peyro M, Soheilypour M, Ghavami A, Mofrad MR. Peyro M, et al. PLoS One. 2015 Dec 11;10(12):e0143745. doi: 10.1371/journal.pone.0143745. eCollection 2015. PLoS One. 2015. PMID: 26658558 Free PMC article. - Thermodynamic characterization of the multivalent interactions underlying rapid and selective translocation through the nuclear pore complex.
Hayama R, Sparks S, Hecht LM, Dutta K, Karp JM, Cabana CM, Rout MP, Cowburn D. Hayama R, et al. J Biol Chem. 2018 Mar 23;293(12):4555-4563. doi: 10.1074/jbc.AC117.001649. Epub 2018 Jan 26. J Biol Chem. 2018. PMID: 29374059 Free PMC article. - Interaction of nucleoporins with nuclear transport receptors: a structural perspective.
Kehlenbach RH, Neumann P, Ficner R, Dickmanns A. Kehlenbach RH, et al. Biol Chem. 2023 May 22;404(8-9):791-805. doi: 10.1515/hsz-2023-0155. Print 2023 Jul 26. Biol Chem. 2023. PMID: 37210735 Review. - Distinct, but not completely separate spatial transport routes in the nuclear pore complex.
Yang W. Yang W. Nucleus. 2013 May-Jun;4(3):166-75. doi: 10.4161/nucl.24874. Epub 2013 May 1. Nucleus. 2013. PMID: 23669120 Free PMC article. Review.
Cited by
- Nuclear transport proteins: structure, function, and disease relevance.
Yang Y, Guo L, Chen L, Gong B, Jia D, Sun Q. Yang Y, et al. Signal Transduct Target Ther. 2023 Nov 10;8(1):425. doi: 10.1038/s41392-023-01649-4. Signal Transduct Target Ther. 2023. PMID: 37945593 Free PMC article. Review. - Simple rules for passive diffusion through the nuclear pore complex.
Timney BL, Raveh B, Mironska R, Trivedi JM, Kim SJ, Russel D, Wente SR, Sali A, Rout MP. Timney BL, et al. J Cell Biol. 2016 Oct 10;215(1):57-76. doi: 10.1083/jcb.201601004. Epub 2016 Oct 3. J Cell Biol. 2016. PMID: 27697925 Free PMC article. - Design principles of selective transport through biopolymer barriers.
Maguire L, Stefferson M, Betterton MD, Hough LE. Maguire L, et al. Phys Rev E. 2019 Oct;100(4-1):042414. doi: 10.1103/PhysRevE.100.042414. Phys Rev E. 2019. PMID: 31770897 Free PMC article. - Free energy calculations shed light on the nuclear pore complex's selective barrier nature.
Matsuda A, Mofrad MRK. Matsuda A, et al. Biophys J. 2021 Sep 7;120(17):3628-3640. doi: 10.1016/j.bpj.2021.07.025. Epub 2021 Jul 31. Biophys J. 2021. PMID: 34339633 Free PMC article. - Nucleoporins' exclusive amino acid sequence features regulate their transient interaction with and selectivity of cargo complexes in the nuclear pore.
Peyro M, Dickson AM, Mofrad MRK. Peyro M, et al. Mol Biol Cell. 2021 Nov 1;32(21):ar31. doi: 10.1091/mbc.E21-04-0161. Epub 2021 Sep 2. Mol Biol Cell. 2021. PMID: 34473567 Free PMC article.
References
- Simon DN, Rout MP. Cancer and the nuclear pore complex. Adv Exp Med Biol. 2014;773:285–307. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM083960/GM/NIGMS NIH HHS/United States
- S10 OD016305/OD/NIH HHS/United States
- R01 GM112108/GM/NIGMS NIH HHS/United States
- P41 GM103712/GM/NIGMS NIH HHS/United States
- R01 GM117212/GM/NIGMS NIH HHS/United States
- T32 GM007288/GM/NIGMS NIH HHS/United States
- P41 GM109824/GM/NIGMS NIH HHS/United States
- U01 GM098256/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous