Untangling the brain's neuroinflammatory and neurodegenerative transcriptional responses - PubMed (original) (raw)
doi: 10.1038/ncomms11295.
Brad A Friedman 2, Jessica L Larson 2, Benjamin E Lauffer 1, Leonard D Goldstein 2 3, Laurie L Appling 4, Jovencio Borneo 4, Chungkee Poon 4, Terence Ho 4, Fang Cai 5, Pascal Steiner 1, Marcel P van der Brug 5, Zora Modrusan 3, Joshua S Kaminker 2, David V Hansen 1
Affiliations
- PMID: 27097852
- PMCID: PMC4844685
- DOI: 10.1038/ncomms11295
Untangling the brain's neuroinflammatory and neurodegenerative transcriptional responses
Karpagam Srinivasan et al. Nat Commun. 2016.
Abstract
A common approach to understanding neurodegenerative disease is comparing gene expression in diseased versus healthy tissues. We illustrate that expression profiles derived from whole tissue RNA highly reflect the degenerating tissues' altered cellular composition, not necessarily transcriptional regulation. To accurately understand transcriptional changes that accompany neuropathology, we acutely purify neurons, astrocytes and microglia from single adult mouse brains and analyse their transcriptomes by RNA sequencing. Using peripheral endotoxemia to establish the method, we reveal highly specific transcriptional responses and altered RNA processing in each cell type, with Tnfr1 required for the astrocytic response. Extending the method to an Alzheimer's disease model, we confirm that transcriptomic changes observed in whole tissue are driven primarily by cell type composition, not transcriptional regulation, and identify hundreds of cell type-specific changes undetected in whole tissue RNA. Applying similar methods to additional models and patient tissues will transform our understanding of aberrant gene expression in neurological disease.
Conflict of interest statement
The authors all are/were employees of Genentech Inc. and declare no competing financial interests.
Figures
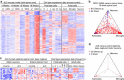
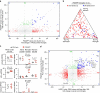
Figure 1. Mapping dementia-related gene expression profiles to CNS cell types.
(a) Genes differentially expressed by microarray analysis (fold change ⩾2, adjusted _P_≤0.05) in GRN mutant FTD patient cortex versus controls (GSE13162, imported) were analysed for expression in cerebrocortical cell types from normal postnatal (GSE52564, imported) or adult (GSE75246, this study) mouse brains. Each gene's expression was _Z_-score normalized separately within each of the three data sets (colour range: −4.10≤Z≤5.23). Rows (genes) were clustered hierarchically. Columns (RNA samples) were sorted by sample metadata. (b) The same genes with higher (red) or lower (blue) expression in GRN mutant FTD cortex are plotted in this triangle. A gene's proximity to a corner represents its degree of preferential expression in the indicated cell type from normal adult mouse cortex (see the Methods for details). (c,d) Similar to a,b, this heat map (colour range: −2.20≤Z≤3.47) and triangle plot analyse cell type expression for genes differentially expressed by microarray in bulk cerebral cortex from the PS2APP Alzheimer's model versus wild-type mice at 13 months of age (GSE74995, this study). a/astro., astrocytes; c, control; e, endothelial cells; m/micro., microglia; mo, myelinating oligodendrocytes; n, neurons; nfo, newly formed oligodendrocytes; opc, oligodendrocyte progenitor cells; wt, wild type.
Figure 2. Mapping ALS-related gene expression profiles to CNS cell types.
(a,b) Similar to Fig. 1a,b, this heat map (colour range: −3.34≤Z≤3.68) and triangle plot analyse cell type expression for genes differentially expressed in bulk spinal cords from the SOD1 G93A ALS model versus wild-type mice at 18 weeks of age (GSE18597, imported). (c,d) Similar to Fig. 1a,b, this heat map (colour range: −2.75≤Z≤3.14) and triangle plot analyse cell type expression for genes differentially expressed in laser-captured motor neuron samples from ALS patient spinal cords versus controls (GSE18920, imported). Expression data from anterior horn material remaining after motor neuron excision are also shown. a/astro., astrocytes; Cx, cortex; e, endothelial cells; m/micro., microglia; mo, myelinating oligodendrocytes; n, neurons; nfo, newly formed oligodendrocytes; opc, oligodendrocyte progenitor cells; wt, wild type.
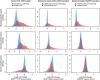
Figure 3. Biased expression of cell type-specific genes in neurodegenerative tissues.
Genes specifically expressed by microglia, astrocytes or neurons were selected as those with ⩾20-fold enrichment compared with the other two cell types when isolated from normal adult mouse cortex and analysed by RNA-seq. For the subsets of these genes or their human homologues that were probed by microarray, distributions are shown for fold-changes in bulk cortex from FTD patients, bulk spinal cord from murine ALS model or bulk cortex from murine AD model, relative to controls, in unaffected (blue) and affected (red) regions and times. The shift of a red distribution to the right is consistent with higher content of that cell type in the affected tissue (for example, microglia in AD model and astrocytes in ALS model), whereas a shift to the left suggests a depletion of that cell type (for example, neurons in FTD cortex).
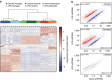
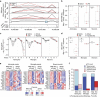
Figure 4. Antibody-based FACS and RNA-Seq for CNS cell types from adult brain tissue.
(a) Left panel shows forward/side scatter (FSC/SSC) of dissociated cortical tissue. DAPI+ events (‘cells') were gated to select for nuclei-containing singlets (second panel), and neurons and microglia were isolated as two distinct populations corresponding to NeuN+CD11b– and NeuN−CD11b+ cells, respectively (third panel). The NeuN− population was further used to isolate GFAP+ astrocytes (last panel). (b) RNA-Seq data confirmed that NeuN+ sorting enriched for cells expressing neuronal markers, GFAP+ sorting enriched for cells expressing astrocytic markers, and CD11b+ sorting enriched for cells expressing microglial markers (_n_=5 animals; one astrocyte sample was excluded from the analysis due to evidence of neuronal contamination). Bars represent mean±s.d. (Prism).
Figure 5. Genome-wide, cell type-specific expression profiles of the brain's endotoxic transcriptional response.
(a) We performed unsupervised clustering of sorted brain cell RNA-Seq data from saline- and LPS-injected mice (_n_=5 animals per treatment) using the 2.5% most variably expressed genes across all samples. The samples segregated into distinct clusters determined primarily by lineage and cell type. Microglia and astrocyte samples further segregated according to treatment. (One astrocyte sample from each group was excluded from the analysis due to evidence of neuronal contamination.) (b) Genome-wide ‘two-way' plots of average gene expression levels in microglia, astrocytes or neurons isolated from LPS-injected versus saline-injected mice. Genes whose average expression increased (red) or decreased (blue) at least fourfold (adjusted _P_≤0.05, Wald test, DESeq2) are plotted.
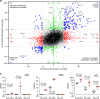
Figure 6. Distinct transcriptional responses to endotoxemia in microglia and astrocytes.
(a) Genome-wide ‘four-way' plot for every gene's LPS-induced fold change (FC) in astrocyte expression versus LPS-induced FC in microglial expression. Each coloured dot represents a gene that met the cutoffs of FC⩾4, adjusted _P_≤0.05 in microglia only (red), in astrocytes only (green) or in both cell types (blue). Higher induction in terms of FC should not be conflated with absolute expression levels (see examples in b). Genes below an expression threshold (see the Methods for details) in a cell type are plotted as FC=1 for that cell type. (b) Individual expression plots of RNA-Seq data for Irak3, Slfn8 and Mt2 genes in each cell type from saline- and LPS-injected mice (_n_=5 animals per treatment), with FC and adjusted _P_-values shown (Wald test, DESeq2). Bars represent mean±s.d. (calculated by Prism).
Figure 7. Endotoxemia-induced RNA splicing alterations in CNS cell types.
(a) RNA-Seq junction reads for example event in Sltm gene. Peaks represent detected splice junctions with heights indicating average size factor-normalized coverage within each treatment group. The skipping junction (longer peak) is detected in all three cell types from LPS-treated mice but very low in the vehicle group. Three annotated Sltm exons (cassette exon in blue) and qPCR primers are shown below. Grey bar for the skipping assay indicates a junction-spanning primer. (b) RNA-Seq reads from same Sltm event summarized as variant frequencies (vFreq) between 0 (complete skipping) and 1 (complete inclusion). The single samples in the microglia and neuron LPS-treated groups that maintained a high level of inclusion were from the same animal, ‘LPS6'. (c) −Δ_C_t qPCR results for example event in Sltm. Constitutive (C), skipping (S) and inclusion (I) assays were analysed in a replication cohort of four animals per treatment. Lines connect results for individual animals. Greater −Δ_C_t values indicate higher abundance of the corresponding Sltm isoform. Higher ‘S' values, relative to ‘I' values, in LPS samples indicate more skipping. (d) ΔΔ_C_t qPCR results, defined as (−Δ_C_tinclusion)−(−Δ_C_tskip), for same Sltm event confirm increased exon skipping upon LPS treatment. Lower ΔΔ_C_t values indicate more skipping, as seen in the LPS-treated samples. (e) Differential splicing events identified by RNA-Seq and tested by RT–qPCR. Variant frequency (vFreq) and ΔΔ_C_t for each event were Z-score normalized within each cell type. White colour indicates average inclusion for that event and cell type; red or blue indicate above average inclusion or skipping, respectively. (f) Summary of qPCR results for validating the LPS-induced RNA splicing alterations identified by RNA-Seq. qPCR results are summarized based on average ΔΔ_C_t for direction of change and significant (sig.) or not based on adjusted _t_-test _P_≤0.05. Cont., control.
Figure 8. TNF receptor and cell type specificity in CNS endotoxic response.
(a) RNA-Seq data showing expression levels of TNF, Tnfr1 and Tnfr2 in microglia and astrocytes, with LPS-induced fold changes (FCs) and adjusted _P_-values shown (Wald test, DESeq2; _n_=5 animals per treatment). Neuronal expression of Tnf receptors was negligible. Bars represent mean±s.d. (Prism). (b) Wild-type and TNFR knockout colony mates were injected with saline or LPS (_n_=5 per treatment and genotype). Cell types were purified 1 day post injection and tested by RT–qPCR for transcriptional responses, normalized against averaged _C_t values for Gapdh and Rpl37a. Comparing LPS-induced changes (ΔΔ_C_t=average−Δ_C_tLPS minus average −Δ_C_tsaline) between wild-type and knockout cell types revealed a strong dependence on Tnfr1 for the much of the astrocytic response (top left plot). Tnfr1−/− microglia responded normally overall (top right plot), although a few genes (see Mt2) displayed Tnfr1 dependence. The absence of Tnfr2 had no strong effect on any cell type (bottom plots).
Figure 9. Gliosis entirely accounts for amyloid-driven changes in whole tissue RNA profiles.
(a) Heat map of RNA-Seq data relating the genes with ⩾2-fold change (FC; adjusted _P_≤0.05) in 13-month-old PS2APP whole cortex (GSE75357, this study) to expression data for astrocytes, microglia and neurons acutely isolated from aged transgenic (Tg) and wild-type (wt) mice (GSE75431, this study). Most genes with increased abundance in PS2APP cortex were preferentially expressed by microglia, but only a fraction of these were transcriptionally upregulated in microglia. Expression was _Z_-score normalized for each gene, separately within each of the two data sets (colour range: −2.03≤Z≤4.00). One astrocyte sample from PS2APP mice was excluded from the analysis due to evidence of cell type contamination. (b,c) Expression patterns of indicated genes analysed by RNA-Seq for whole cortex and for sorted cells, from PS2APP versus non-transgenic cortices, with FC and adjusted _P_-values (Wald test, DESeq2; _n_=5 animals per genotype) shown and bars representing mean±s.d. (Prism). Note: Absolute nRPKM values between whole cortex and sorted cell studies should not be directly compared, due to different library preparation methods.
Figure 10. Analysis of amyloid-driven changes in microglial gene expression.
(a) Genome-wide ‘four-way' plot comparing differential gene expression (fold change (FC) ⩾2, adjusted _P_≤0.5) observed in PS2APP whole cortex versus that observed in purified PS2APP microglia (each compared with non-transgenic). Each point represents a single gene, with only genes meeting the cutoffs in either study being plotted. Red, green and blue points met the cutoffs in sorted microglia only, whole cortex only or in both analyses, respectively. Grey indicates the distribution of FC values for all genes. (b) In this triangle plot, the 249 genes differentially expressed in PS2APP microglia (FC ⩾2, adjusted _P_≤0.05) are plotted to show their degree of preferential expression in neurons, astrocytes or microglia from normal, non-transgenic (non-Tg) cortex. (c) Specific examples of amyloid-driven changes in microglial gene expression that were undetected or muted in whole tissue expression profiles, with FC and adjusted _P_-values (Wald test, DESeq2; _n_=5 animals per genotype) shown and bars representing mean±s.d. (Prism). Note: Absolute nRPKM values between whole cortex and sorted cell studies should not be directly compared, because of different library preparation methods. (d) Similar to a, except this ‘four-way' plot compares genes differentially expressed in microglia from PS2APP Alzheimer's model (this study) with those observed in microglia from the hSOD1mut ALS model (GSE43366, imported).
Similar articles
- Endotoxemia-induced cytokine-mediated responses of hippocampal astrocytes transmitted by cells of the brain-immune interface.
Hasegawa-Ishii S, Inaba M, Umegaki H, Unno K, Wakabayashi K, Shimada A. Hasegawa-Ishii S, et al. Sci Rep. 2016 May 5;6:25457. doi: 10.1038/srep25457. Sci Rep. 2016. PMID: 27149601 Free PMC article. - Alzheimer's Patient Microglia Exhibit Enhanced Aging and Unique Transcriptional Activation.
Srinivasan K, Friedman BA, Etxeberria A, Huntley MA, van der Brug MP, Foreman O, Paw JS, Modrusan Z, Beach TG, Serrano GE, Hansen DV. Srinivasan K, et al. Cell Rep. 2020 Jun 30;31(13):107843. doi: 10.1016/j.celrep.2020.107843. Cell Rep. 2020. PMID: 32610143 Free PMC article. - S100 beta and serotonin: a possible astrocytic-neuronal link to neuropathology of Alzheimer's disease.
Azmitia EC, Griffin WS, Marshak DR, Van Eldik LJ, Whitaker-Azmitia PM. Azmitia EC, et al. Prog Brain Res. 1992;94:459-73. Prog Brain Res. 1992. PMID: 1287730 Review. No abstract available. - A core transcriptional signature of human microglia: Derivation and utility in describing region-dependent alterations associated with Alzheimer's disease.
Patir A, Shih B, McColl BW, Freeman TC. Patir A, et al. Glia. 2019 Jul;67(7):1240-1253. doi: 10.1002/glia.23572. Epub 2019 Feb 13. Glia. 2019. PMID: 30758077 Review.
Cited by
- Neuroinflammatory gene expression profiles of reactive glia in the substantia nigra suggest a multidimensional immune response to alpha synuclein inclusions.
Stoll AC, Kemp CJ, Patterson JR, Howe JW, Steece-Collier K, Luk KC, Sortwell CE, Benskey MJ. Stoll AC, et al. Neurobiol Dis. 2024 Feb;191:106411. doi: 10.1016/j.nbd.2024.106411. Epub 2024 Jan 14. Neurobiol Dis. 2024. PMID: 38228253 Free PMC article. - Inhibition of the CEBPβ-NFκB interaction by nanocarrier-packaged Carnosic acid ameliorates glia-mediated neuroinflammation and improves cognitive function in an Alzheimer's disease model.
Yi-Bin W, Xiang L, Bing Y, Qi Z, Fei-Tong J, Minghong W, Xiangxiang Z, Le K, Yan L, Ping S, Yufei G, Ye X, Chun-Yan W. Yi-Bin W, et al. Cell Death Dis. 2022 Apr 7;13(4):318. doi: 10.1038/s41419-022-04765-1. Cell Death Dis. 2022. PMID: 35393391 Free PMC article. - Endometrial organoids derived from Mayer-Rokitansky-Küster-Hauser syndrome patients provide insights into disease-causing pathways.
Brucker SY, Hentrich T, Schulze-Hentrich JM, Pietzsch M, Wajngarten N, Singh AR, Rall K, Koch A. Brucker SY, et al. Dis Model Mech. 2022 May 1;15(5):dmm049379. doi: 10.1242/dmm.049379. Epub 2022 May 10. Dis Model Mech. 2022. PMID: 35394036 Free PMC article. - Paired Immunoglobulin-like Type 2 Receptor Alpha G78R variant alters ligand binding and confers protection to Alzheimer's disease.
Rathore N, Ramani SR, Pantua H, Payandeh J, Bhangale T, Wuster A, Kapoor M, Sun Y, Kapadia SB, Gonzalez L, Zarrin AA, Goate A, Hansen DV, Behrens TW, Graham RR. Rathore N, et al. PLoS Genet. 2018 Nov 2;14(11):e1007427. doi: 10.1371/journal.pgen.1007427. eCollection 2018 Nov. PLoS Genet. 2018. PMID: 30388101 Free PMC article. - A Model of Discovery: The Role of Imaging Established and Emerging Non-mammalian Models in Neuroscience.
Haynes EM, Ulland TK, Eliceiri KW. Haynes EM, et al. Front Mol Neurosci. 2022 Apr 14;15:867010. doi: 10.3389/fnmol.2022.867010. eCollection 2022. Front Mol Neurosci. 2022. PMID: 35493325 Free PMC article. Review.
References
- Cardona A. E., Huang D., Sasse M. E. & Ransohoff R. M. Isolation of murine microglial cells for RNA analysis or flow cytometry. Nat. Protoc. 1, 1947–1951 (2006) . - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases