CoinFold: a web server for protein contact prediction and contact-assisted protein folding - PubMed (original) (raw)
. 2016 Jul 8;44(W1):W361-6.
doi: 10.1093/nar/gkw307. Epub 2016 Apr 25.
Affiliations
- PMID: 27112569
- PMCID: PMC4987891
- DOI: 10.1093/nar/gkw307
CoinFold: a web server for protein contact prediction and contact-assisted protein folding
Sheng Wang et al. Nucleic Acids Res. 2016.
Abstract
CoinFold (http://raptorx2.uchicago.edu/ContactMap/) is a web server for protein contact prediction and contact-assisted de novo structure prediction. CoinFold predicts contacts by integrating joint multi-family evolutionary coupling (EC) analysis and supervised machine learning. This joint EC analysis is unique in that it not only uses residue coevolution information in the target protein family, but also that in the related families which may have divergent sequences but similar folds. The supervised learning further improves contact prediction accuracy by making use of sequence profile, contact (distance) potential and other information. Finally, this server predicts tertiary structure of a sequence by feeding its predicted contacts and secondary structure to the CNS suite. Tested on the CASP and CAMEO targets, this server shows significant advantages over existing ones of similar category in both contact and tertiary structure prediction.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
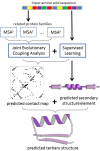
Figure 1.
Illustration of CoinFold workflow. Given an input protein sequence, CoinFold uses HHblits (22) and HHpred (23) to generate sequence profile and search for related protein families. Then CoinFold conducts joint evolutionary coupling analysis and supervised prediction of both contacts and secondary structure. Finally, CoinFold predicts 3D models using CNS.
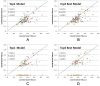
Figure 2.
TMscore comparison of the top models generated by CoinFold, ConFold, and EVfold. The top 1 and the best (in terms of TMscore) of top 5 models are evaluated. The 36 CASP10 targets, 49 from CASP11 targets and 47 CAMEO targets are shown in red square, blue diamond, and green triangle, respectively. (A and B) Head-to-head comparison of Top1 and Top5 best models by CoinFold (in X-axis) and ConFold (in Y-axis). (C and D) Head-to-head comparison of Top1 and Top5 best models by CoinFold (in X-axis) and EVfold (in Y-axis).
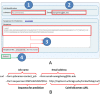
Figure 3.
CoinFold server job submission. (A) The web interface for job submission has fields for job name (1), optional user email address (2), and sequences to be submitted (3). The sequences shall be in FASTA format and can also be submitted in a file (3). User can also click on the example link to see an example. Submit a job by clicking on the submit button (4). (B) An example for submission by a publicly available program Curl. Only ‘sequences’ and the submission URL (shown in underlines) are required and the others are optional. A job URL will be returned on screen after submission.
Figure 4.
CoinFold server result page. The left part shows the predicted contact map (1), where the predicted score is displayed in greyscale, with a higher score represented by a darker color. The middle part shows the job status (the submitted, scheduled, and finished time) (2), as well as two downloading buttons for the predicted contact map (3) and 5 predicted 3D models (4). The right part contains a button to view alternative 3D models (5), a display bar for showing rank and score of the selected 3D model (6) and visualization of the selected 3D model (7).
Similar articles
- Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model.
Wang S, Sun S, Li Z, Zhang R, Xu J. Wang S, et al. PLoS Comput Biol. 2017 Jan 5;13(1):e1005324. doi: 10.1371/journal.pcbi.1005324. eCollection 2017 Jan. PLoS Comput Biol. 2017. PMID: 28056090 Free PMC article. - Protein contact prediction by integrating joint evolutionary coupling analysis and supervised learning.
Ma J, Wang S, Wang Z, Xu J. Ma J, et al. Bioinformatics. 2015 Nov 1;31(21):3506-13. doi: 10.1093/bioinformatics/btv472. Epub 2015 Aug 14. Bioinformatics. 2015. PMID: 26275894 Free PMC article. - RaptorX-Property: a web server for protein structure property prediction.
Wang S, Li W, Liu S, Xu J. Wang S, et al. Nucleic Acids Res. 2016 Jul 8;44(W1):W430-5. doi: 10.1093/nar/gkw306. Epub 2016 Apr 25. Nucleic Acids Res. 2016. PMID: 27112573 Free PMC article. - State-of-the-art web services for de novo protein structure prediction.
Abriata LA, Dal Peraro M. Abriata LA, et al. Brief Bioinform. 2021 May 20;22(3):bbaa139. doi: 10.1093/bib/bbaa139. Brief Bioinform. 2021. PMID: 34020540 Review. - Prediction of protein contacts from correlated sequence substitutions.
Sadowski MI, Taylor WR. Sadowski MI, et al. Sci Prog. 2013;96(Pt 1):33-42. doi: 10.3184/003685013X13612883013639. Sci Prog. 2013. PMID: 23738436 Free PMC article. Review.
Cited by
- Three-Dimensional Graph Matching to Identify Secondary Structure Correspondence of Medium-Resolution Cryo-EM Density Maps.
Behkamal B, Naghibzadeh M, Saberi MR, Tehranizadeh ZA, Pagnani A, Al Nasr K. Behkamal B, et al. Biomolecules. 2021 Nov 26;11(12):1773. doi: 10.3390/biom11121773. Biomolecules. 2021. PMID: 34944417 Free PMC article. - A single-model quality assessment method for poor quality protein structure.
Ouyang J, Huang N, Jiang Y. Ouyang J, et al. BMC Bioinformatics. 2020 Apr 25;21(1):157. doi: 10.1186/s12859-020-3499-5. BMC Bioinformatics. 2020. PMID: 32334508 Free PMC article. - Evaluating the significance of contact maps in low-homology protein modeling using contact-assisted threading.
Bhattacharya S, Bhattacharya D. Bhattacharya S, et al. Sci Rep. 2020 Feb 19;10(1):2908. doi: 10.1038/s41598-020-59834-2. Sci Rep. 2020. PMID: 32076047 Free PMC article. - Design of novel cyanovirin-N variants by modulation of binding dynamics through distal mutations.
Kazan IC, Sharma P, Rahman MI, Bobkov A, Fromme R, Ghirlanda G, Ozkan SB. Kazan IC, et al. Elife. 2022 Dec 6;11:e67474. doi: 10.7554/eLife.67474. Elife. 2022. PMID: 36472898 Free PMC article. - Contact-Assisted Threading in Low-Homology Protein Modeling.
Bhattacharya S, Roche R, Shuvo MH, Moussad B, Bhattacharya D. Bhattacharya S, et al. Methods Mol Biol. 2023;2627:41-59. doi: 10.1007/978-1-0716-2974-1_3. Methods Mol Biol. 2023. PMID: 36959441 Free PMC article.
References
- de Juan D., Pazos F., Valencia A. Emerging methods in protein co-evolution. Nat. Rev. Genet. 2013;14:249–261. - PubMed
- Göbel U., Sander C., Schneider R., Valencia A. Correlated mutations and residue contacts in proteins. Proteins: Struct. Funct. Bioinform. 1994;18:309–317. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources