The Sus operon: a model system for starch uptake by the human gut Bacteroidetes - PubMed (original) (raw)
Review
The Sus operon: a model system for starch uptake by the human gut Bacteroidetes
Matthew H Foley et al. Cell Mol Life Sci. 2016 Jul.
Abstract
Resident bacteria in the densely populated human intestinal tract must efficiently compete for carbohydrate nutrition. The Bacteroidetes, a dominant bacterial phylum in the mammalian gut, encode a plethora of discrete polysaccharide utilization loci (PULs) that are selectively activated to facilitate glycan capture at the cell surface. The most well-studied PUL-encoded glycan-uptake system is the starch utilization system (Sus) of Bacteroides thetaiotaomicron. The Sus includes the requisite proteins for binding and degrading starch at the surface of the cell preceding oligosaccharide transport across the outer membrane for further depolymerization to glucose in the periplasm. All mammalian gut Bacteroidetes possess analogous Sus-like systems that target numerous diverse glycans. In this review, we discuss what is known about the eight Sus proteins of B. thetaiotaomicron that define the Sus-like paradigm of nutrient acquisition that is exclusive to the Gram-negative Bacteroidetes. We emphasize the well-characterized outer membrane proteins SusDEF and the α-amylase SusG, each of which have unique structural features that allow them to interact with starch on the cell surface. Despite the apparent redundancy in starch-binding sites among these proteins, each has a distinct role during starch catabolism. Additionally, we consider what is known about how these proteins dynamically interact and cooperate in the membrane and propose a model for the formation of the Sus outer membrane complex.
Keywords: Bacteroides thetaiotaomicron; Gut microbiota; Starch; Starch utilization system; Sus.
Figures
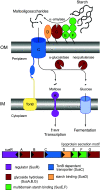
Fig. 1
Overview of the starch utilization system (Sus) in B. thetaiotaomicron. The sus locus is transcribed from two divergent promoters. Transcription of susR occurs independently from the rest of the locus, which allows the inner membrane-spanning protein SusR to sense the disaccharide inducer, maltose, in the periplasm and subsequently drive the transcription of susABCDEFG. Starch is bound to the surface of the cell by the starch-binding outer membrane lipoproteins SusDEF. Subsequent hydrolysis by a similarly membrane-tethered α-amylase, SusG, generates oligosaccharides small enough to transit through the TonB-dependent transporter. Once in the periplasm, SusA and SusB, a neopullulanase and α-glucosidase, respectively, process oligosaccharides into glucose. The monosaccharide is then shuttled into the cytoplasm by an unknown transporter. The stoichiometry and assembly of the Sus is unknown
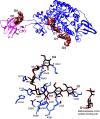
Fig. 2
Molecular structure of SusD with maltooligosaccharides. a Superposition of SusD (blue, PDB 3CK9) with bound maltoheptaose (blue sticks) and the SusD homolog BT1043 (gray, PDB 3EHN) that targets mucosal glycans with bound _N_-acetyllactosamine (black sticks). The conservation of the eight tetratricopeptide repeat helices is highlighted in darker colors for both proteins. The RMSD for these proteins is 2.8 Å over 324 aligned residues (12.6 % sequence identity). b Superposition of the structure of SusD with bound maltoheptaose (blue) and maltotriose (pink), highlighting the plasticity within the binding site. Residues that move upon binding of a longer α-glucan are in bold print. c SusD crystallized with α-cyclodextrin revealed protein dimerization. The affinity of starch-binding to the cell surface may be enhanced by an avidity effect, whereby multiple SusD proteins cooperate to bind the polymer
Fig. 3
SusG is an amylase with a unique CBM insertion. a Structure of the catalytically inactive mutant of SusG D498 N (PDB 3K8L) with bound maltoheptaose. CBM58 (residues 216–335) is highlighted in pink, and maltooligosaccharides bound at CBM58, the active site, and the surface starch-binding site are depicted as spheres. The orientation of the oligosaccharide from the nonreducing end (O4) to reducing end (O1) is indicated. b Close-up view of the active site in the catalytically inactive mutant of SusG D498 N (PDB 3K8L) with bound maltoheptaose. Hydrogen-bonding interactions (≤3.5 Å) are depicted as dashed lines, and Glc residues are labeled from the non-reducing to reducing end
Fig. 4
Structures of the SusE and SusF proteins. a Structure of SusE with bound α-cyclodextrin (PDB 4FEM), with the starch-binding domains Eb and Ec in different colors. Proline residues between the domains are highlighted as spheres. b Structure of SusF with bound maltoheptaose (PDB 4FE9), with the starch-binding domains Fa, Fb, and Fc in different colors. Proline residues between domains are highlighted as spheres. c Overlay of the Eb/Ec and Fb/Fc domains of SusE and SusF, colored as in panels a and b. d Superposition of the Eb domain (blue), Fb domain (pink) and the X25 domain (black, residues 161-270 of PDB 2WAN) from the Bacillus acidopullulyticus pullulanase [76]. e Close-up of the starch-binding sites in Eb and Fb from the overlay in panel d, demonstrating that these residues are conserved within the X25 module of the pullulanase (PDB 2WAN). Residues and labels are colored as in panel d, and the portion of the α-cyclodextrin bound to Eb is displayed as transparent orange and red sticks. f Overlay of the two positions that maltoheptaose occupied at the Ec binding site of SusE (PDB 4FCH), demonstrating how a longer single-helical stretch of amylose could be accommodated
Fig. 5
Sus protein structures and model of dynamic assembly. Ribbon diagrams of the crystal structures and homology models for the seven Sus proteins involved in starch utilization in B. thetaiotaomicron, colored as in Fig. 1. The flexible amino acid sequences that link SusDEFG to the membrane are depicted as a black wavy lines, as these residues were not resolved in the crystal structures. SusG (PDB 3K8L) dynamically interacts with SusD (PDB 3CK9) and SusC (ITASSER structure prediction [85, 86]). SusE (PDB 4FEM; ITASSER prediction of the N-terminal domain, with modeling of the linker sequence) and SusF (PDB 4FE9) may directly interact with each other and with the SusCD complex, as suggested by Salyers [27]. Maltooligosaccharides are transported through the SusC TonB-dependent transporter where they are further hydrolyzed to glucose by the action of SusB (PDB 2JKA) and SusA (model from Modpipe [87] using template PDB 3DHU). Maltooligosaccharides and glucose are depicted as black and red sticks
Fig. 6
Sus operon structure across Bacteroides species. Selected sus operons from the type strains of several human gut Bacteroides species are displayed, identified via conservation of the operon structure surrounding a susG homolog. Numbers displayed above each gene indicate the percent identity of the encoded protein and in parentheses the percent coverage of the match to the homologous protein from B. thetaiotaomicron. For example, 63 % (100 %) above the susC homolog from B. ovatus indicates that the SusC homolog from B. ovatus is 63 % identical to the B. thetaiotaomicron SusC and that this match covers 100 % of the protein sequence
Similar articles
- Superresolution imaging captures carbohydrate utilization dynamics in human gut symbionts.
Karunatilaka KS, Cameron EA, Martens EC, Koropatkin NM, Biteen JS. Karunatilaka KS, et al. mBio. 2014 Nov 11;5(6):e02172. doi: 10.1128/mBio.02172-14. mBio. 2014. PMID: 25389179 Free PMC article. - Multifunctional nutrient-binding proteins adapt human symbiotic bacteria for glycan competition in the gut by separately promoting enhanced sensing and catalysis.
Cameron EA, Kwiatkowski KJ, Lee BH, Hamaker BR, Koropatkin NM, Martens EC. Cameron EA, et al. mBio. 2014 Sep 9;5(5):e01441-14. doi: 10.1128/mBio.01441-14. mBio. 2014. PMID: 25205092 Free PMC article. - SusE facilitates starch uptake independent of starch binding in B. thetaiotaomicron.
Foley MH, Martens EC, Koropatkin NM. Foley MH, et al. Mol Microbiol. 2018 Jun;108(5):551-566. doi: 10.1111/mmi.13949. Epub 2018 Apr 14. Mol Microbiol. 2018. PMID: 29528148 Free PMC article. - Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm.
Martens EC, Koropatkin NM, Smith TJ, Gordon JI. Martens EC, et al. J Biol Chem. 2009 Sep 11;284(37):24673-7. doi: 10.1074/jbc.R109.022848. Epub 2009 Jun 24. J Biol Chem. 2009. PMID: 19553672 Free PMC article. Review. - TonB-dependent transporters in the Bacteroidetes: Unique domain structures and potential functions.
Pollet RM, Martin LM, Koropatkin NM. Pollet RM, et al. Mol Microbiol. 2021 Mar;115(3):490-501. doi: 10.1111/mmi.14683. Epub 2021 Feb 3. Mol Microbiol. 2021. PMID: 33448497 Review.
Cited by
- Wheat supplement with buckwheat affect gut microbiome composition and circulate short-chain fatty acids.
Yao D, Yu Q, Xu L, Su T, Ma L, Wang X, Wu M, Li Z, Zhang D, Wang C. Yao D, et al. Front Nutr. 2022 Sep 6;9:952738. doi: 10.3389/fnut.2022.952738. eCollection 2022. Front Nutr. 2022. PMID: 36147303 Free PMC article. - Roles of the Cell Surface Architecture of Bacteroides and Bifidobacterium in the Gut Colonization.
Nishiyama K, Yokoi T, Sugiyama M, Osawa R, Mukai T, Okada N. Nishiyama K, et al. Front Microbiol. 2021 Oct 14;12:754819. doi: 10.3389/fmicb.2021.754819. eCollection 2021. Front Microbiol. 2021. PMID: 34721360 Free PMC article. Review. - Cultivable, Host-Specific Bacteroidetes Symbionts Exhibit Diverse Polysaccharolytic Strategies.
Vera-Ponce de León A, Jahnes BC, Duan J, Camuy-Vélez LA, Sabree ZL. Vera-Ponce de León A, et al. Appl Environ Microbiol. 2020 Apr 1;86(8):e00091-20. doi: 10.1128/AEM.00091-20. Print 2020 Apr 1. Appl Environ Microbiol. 2020. PMID: 32060023 Free PMC article. - Resistant starch, microbiome, and precision modulation.
Dobranowski PA, Stintzi A. Dobranowski PA, et al. Gut Microbes. 2021 Jan-Dec;13(1):1926842. doi: 10.1080/19490976.2021.1926842. Gut Microbes. 2021. PMID: 34275431 Free PMC article. Review. - Three marine species of the genus Fulvivirga, rich sources of carbohydrate-active enzymes degrading alginate, chitin, laminarin, starch, and xylan.
Nguyen TTH, Vuong TQ, Han HL, Li Z, Lee YJ, Ko J, Nedashkovskaya OI, Kim SG. Nguyen TTH, et al. Sci Rep. 2023 Apr 18;13(1):6301. doi: 10.1038/s41598-023-33408-4. Sci Rep. 2023. PMID: 37072506 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- P30 DK034933/DK/NIDDK NIH HHS/United States
- R01 GM118475/GM/NIGMS NIH HHS/United States
- T32 GM008353/GM/NIGMS NIH HHS/United States
- T32 GM145304/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources