A Polymorphic Antioxidant Response Element Links NRF2/sMAF Binding to Enhanced MAPT Expression and Reduced Risk of Parkinsonian Disorders - PubMed (original) (raw)
A Polymorphic Antioxidant Response Element Links NRF2/sMAF Binding to Enhanced MAPT Expression and Reduced Risk of Parkinsonian Disorders
Xuting Wang et al. Cell Rep. 2016.
Abstract
The NRF2/sMAF protein complex regulates the oxidative stress response by occupying cis-acting enhancers containing an antioxidant response element (ARE). Integrating genome-wide maps of NRF2/sMAF occupancy with disease-susceptibility loci, we discovered eight polymorphic AREs linked to 14 highly ranked disease-risk SNPs in individuals of European ancestry. Among these SNPs was rs242561, located within a regulatory region of the MAPT gene (encoding microtubule-associated protein Tau). It was consistently occupied by NRF2/sMAF in multiple experiments and its strong-binding allele associated with higher mRNA levels in cell lines and human brain tissue. Induction of MAPT transcription by NRF2 was confirmed using a human neuroblastoma cell line and a Nrf2-deficient mouse model. Most importantly, rs242561 displayed complete linkage disequilibrium with a highly protective allele identified in multiple GWASs of progressive supranuclear palsy, Parkinson's disease, and corticobasal degeneration. These observations suggest a potential role for NRF2/sMAF in tauopathies and a possible role for NRF2 pathway activators in disease prevention.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
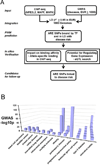
Figure 1
Study design and summary of candidates. (A) Overview of workflow for identifying functional ARE SNPs associated with disease; (B) The significance of GWAS SNPs linked to ARE SNPs.
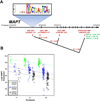
Figure 2
Characteristics of rs242561. (A) location relative to risk SNP for PSP and PD and ChIP-seq signals, (B) position in ARE motif, (C) conservation, (D) allele-specific binding, (E) allele specific transactivation, (F) RT-PCR of MAPT levels in human IMR32 neuroblastoma cells treated with t-BHQ or vehicle, (G) Nrf2 +/+ and Nrf2 −/− mice were exposed to oxidative stress induced by hyperoxia and Mapt gene expression level was measured in mRNA from cerebellum by RT-PCR. See also Figure S3 and Tables S4–S5.
Figure 3
Association of rs242561 genotypes with MAPT mRNA levels in humans. (A) Map of rs242561 and eQTL SNPs with LD correlations (R2) within the MAPT gene; (B) MAPT mRNA levels and genotypes of rs242561 in human brain temporal cortex as measured by Affymetrix Exon array (UKBEC). TT is the stronger binding genotype. See also Figure S3.
Similar articles
- Cis-element architecture of Nrf2-sMaf heterodimer binding sites and its relation to diseases.
Otsuki A, Yamamoto M. Otsuki A, et al. Arch Pharm Res. 2020 Mar;43(3):275-285. doi: 10.1007/s12272-019-01193-2. Epub 2019 Dec 2. Arch Pharm Res. 2020. PMID: 31792803 Review. - Direct and Specific Functional Evaluation of the Nrf2 and MafG Heterodimer by Introducing a Tethered Dimer into Small Maf-Deficient Cells.
Katsuoka F, Otsuki A, Takahashi M, Ito S, Yamamoto M. Katsuoka F, et al. Mol Cell Biol. 2019 Sep 27;39(20):e00273-19. doi: 10.1128/MCB.00273-19. Print 2019 Oct 15. Mol Cell Biol. 2019. PMID: 31383749 Free PMC article. - Gene regulatory effects of disease-associated variation in the NRF2 network.
Lacher SE, Slattery M. Lacher SE, et al. Curr Opin Toxicol. 2016 Dec;1:71-79. doi: 10.1016/j.cotox.2016.09.001. Epub 2016 Sep 28. Curr Opin Toxicol. 2016. PMID: 28203648 Free PMC article. - Unique cistrome defined as CsMBE is strictly required for Nrf2-sMaf heterodimer function in cytoprotection.
Otsuki A, Suzuki M, Katsuoka F, Tsuchida K, Suda H, Morita M, Shimizu R, Yamamoto M. Otsuki A, et al. Free Radic Biol Med. 2016 Feb;91:45-57. doi: 10.1016/j.freeradbiomed.2015.12.005. Epub 2015 Dec 8. Free Radic Biol Med. 2016. PMID: 26677805 - Crosstalk between Nrf2 and Notch signaling.
Wakabayashi N, Chartoumpekis DV, Kensler TW. Wakabayashi N, et al. Free Radic Biol Med. 2015 Nov;88(Pt B):158-167. doi: 10.1016/j.freeradbiomed.2015.05.017. Epub 2015 May 21. Free Radic Biol Med. 2015. PMID: 26003520 Free PMC article. Review.
Cited by
- Dysregulation of a Heme Oxygenase-Synuclein Axis in Parkinson Disease.
Cressatti M, Schipper HM. Cressatti M, et al. NeuroSci. 2022 May 20;3(2):284-299. doi: 10.3390/neurosci3020020. eCollection 2022 Jun. NeuroSci. 2022. PMID: 39483365 Free PMC article. Review. - Whole-genome sequencing analysis reveals new susceptibility loci and structural variants associated with progressive supranuclear palsy.
Wang H, Chang TS, Dombroski BA, Cheng PL, Patil V, Valiente-Banuet L, Farrell K, Mclean C, Molina-Porcel L, Rajput A, De Deyn PP, Le Bastard N, Gearing M, Kaat LD, Van Swieten JC, Dopper E, Ghetti BF, Newell KL, Troakes C, de Yébenes JG, Rábano-Gutierrez A, Meller T, Oertel WH, Respondek G, Stamelou M, Arzberger T, Roeber S, Müller U, Hopfner F, Pastor P, Brice A, Durr A, Le Ber I, Beach TG, Serrano GE, Hazrati LN, Litvan I, Rademakers R, Ross OA, Galasko D, Boxer AL, Miller BL, Seeley WW, Van Deerlin VM, Lee EB, White CL 3rd, Morris H, de Silva R, Crary JF, Goate AM, Friedman JS, Leung YY, Coppola G, Naj AC, Wang LS; P. S. P. genetics study group; Dalgard C, Dickson DW, Höglinger GU, Schellenberg GD, Geschwind DH, Lee WP. Wang H, et al. Mol Neurodegener. 2024 Aug 16;19(1):61. doi: 10.1186/s13024-024-00747-3. Mol Neurodegener. 2024. PMID: 39152475 Free PMC article. - Whole-Genome Sequencing Analysis Reveals New Susceptibility Loci and Structural Variants Associated with Progressive Supranuclear Palsy.
Wang H, Chang TS, Dombroski BA, Cheng PL, Patil V, Valiente-Banuet L, Farrell K, Mclean C, Molina-Porcel L, Rajput A, De Deyn PP, Bastard NL, Gearing M, Kaat LD, Swieten JCV, Dopper E, Ghetti BF, Newell KL, Troakes C, de Yébenes JG, Rábano-Gutierrez A, Meller T, Oertel WH, Respondek G, Stamelou M, Arzberger T, Roeber S, Müller U, Hopfner F, Pastor P, Brice A, Durr A, Ber IL, Beach TG, Serrano GE, Hazrati LN, Litvan I, Rademakers R, Ross OA, Galasko D, Boxer AL, Miller BL, Seeley WW, Deerlin VMV, Lee EB, White CL 3rd, Morris H, de Silva R, Crary JF, Goate AM, Friedman JS, Leung YY, Coppola G, Naj AC, Wang LS; PSP genetics study group; Dickson DW, Höglinger GU, Schellenberg GD, Geschwind DH, Lee WP. Wang H, et al. medRxiv [Preprint]. 2024 Jan 30:2023.12.28.23300612. doi: 10.1101/2023.12.28.23300612. medRxiv. 2024. PMID: 38234807 Free PMC article. Updated. Preprint. - Aging, NRF2, and TAU: A Perfect Match for Neurodegeneration?
Brackhan M, Arribas-Blazquez M, Lastres-Becker I. Brackhan M, et al. Antioxidants (Basel). 2023 Aug 4;12(8):1564. doi: 10.3390/antiox12081564. Antioxidants (Basel). 2023. PMID: 37627559 Free PMC article. Review. - Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Sengoku T, Shiina M, Suzuki K, Hamada K, Sato K, Uchiyama A, Kobayashi S, Oguni A, Itaya H, Kasahara K, Moriwaki H, Watanabe C, Honma T, Okada C, Baba S, Ohta T, Motohashi H, Yamamoto M, Ogata K. Sengoku T, et al. Nucleic Acids Res. 2022 Nov 28;50(21):12543-12557. doi: 10.1093/nar/gkac1102. Nucleic Acids Res. 2022. PMID: 36454022 Free PMC article.
References
- Ballatore C, Lee VM, Trojanowski JQ. Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat Rev Neurosci. 2007;8:663–672. - PubMed
Grants and funding
- Z01 ES046008/Intramural NIH HHS/United States
- Z01 ES046008-18/Intramural NIH HHS/United States
- Z01 ES100475/Intramural NIH HHS/United States
- Z01 ES100475-06/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases