Metagenomics Reveals a Novel Virophage Population in a Tibetan Mountain Lake - PubMed (original) (raw)
Metagenomics Reveals a Novel Virophage Population in a Tibetan Mountain Lake
Seungdae Oh et al. Microbes Environ. 2016.
Abstract
Virophages are parasites of giant viruses that infect eukaryotic organisms and may affect the ecology of inland water ecosystems. Despite the potential ecological impact, limited information is available on the distribution, diversity, and hosts of virophages in ecosystems. Metagenomics revealed that virophages were widely distributed in inland waters with various environmental characteristics including salinity and nutrient availability. A novel virophage population was overrepresented in a planktonic microbial community of the Tibetan mountain lake, Lake Qinghai. Our study identified coccolithophores and coccolithovirus-like phycodnaviruses in the same community, which may serve as eukaryotic and viral hosts of the virophage population, respectively.
Figures
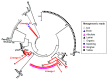
Fig. 1
Occurrence and diversity of virophage populations in diverse terrestrial aquatic environments. The phylogenetic tree was built based on the maximum likelihood method with the Jones-Taylor-Thornton model using MEGA 6.0 (25). Bootstrap support values (higher than 50) from 100 replicates are shown on the nodes of the tree. The three lineages (Lineages I, II, and III) that were not closely related to the previously characterized virophages were highlighted.
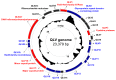
Fig. 2
Circular map of the Qinghai Lake virophage (QLV) genome. Inwards: ORFs of the two DNA strands (red, blue, and black representing core, specific, and others, respectively) and G+C content. Protein-coding genes on the genome were predicted using GeneMark.hmm with the heuristic model (4). The protein-coding genes were functionally annotated by searching the amino acid sequences against the non-redundant protein database (nr) and metagenomic protein (env_nr) database, respectively, using BLASTp with >30% amino acid identity and >50% query length coverage.
Similar articles
- Diversity, evolution, and classification of virophages uncovered through global metagenomics.
Paez-Espino D, Zhou J, Roux S, Nayfach S, Pavlopoulos GA, Schulz F, McMahon KD, Walsh D, Woyke T, Ivanova NN, Eloe-Fadrosh EA, Tringe SG, Kyrpides NC. Paez-Espino D, et al. Microbiome. 2019 Dec 10;7(1):157. doi: 10.1186/s40168-019-0768-5. Microbiome. 2019. PMID: 31823797 Free PMC article. - Novel Cell-Virus-Virophage Tripartite Infection Systems Discovered in the Freshwater Lake Dishui Lake in Shanghai, China.
Xu S, Zhou L, Liang X, Zhou Y, Chen H, Yan S, Wang Y. Xu S, et al. J Virol. 2020 May 18;94(11):e00149-20. doi: 10.1128/JVI.00149-20. Print 2020 May 18. J Virol. 2020. PMID: 32188734 Free PMC article. - The Virophage Family Lavidaviridae.
Fischer MG. Fischer MG. Curr Issues Mol Biol. 2021;40:1-24. doi: 10.21775/cimb.040.001. Epub 2020 Feb 24. Curr Issues Mol Biol. 2021. PMID: 32089519 Review. - Virophages of Giant Viruses: An Update at Eleven.
Mougari S, Sahmi-Bounsiar D, Levasseur A, Colson P, La Scola B. Mougari S, et al. Viruses. 2019 Aug 8;11(8):733. doi: 10.3390/v11080733. Viruses. 2019. PMID: 31398856 Free PMC article. Review.
Cited by
- Diversity, evolution, and classification of virophages uncovered through global metagenomics.
Paez-Espino D, Zhou J, Roux S, Nayfach S, Pavlopoulos GA, Schulz F, McMahon KD, Walsh D, Woyke T, Ivanova NN, Eloe-Fadrosh EA, Tringe SG, Kyrpides NC. Paez-Espino D, et al. Microbiome. 2019 Dec 10;7(1):157. doi: 10.1186/s40168-019-0768-5. Microbiome. 2019. PMID: 31823797 Free PMC article. - The genome of a prasinoviruses-related freshwater virus reveals unusual diversity of phycodnaviruses.
Chen H, Zhang W, Li X, Pan Y, Yan S, Wang Y. Chen H, et al. BMC Genomics. 2018 Jan 15;19(1):49. doi: 10.1186/s12864-018-4432-4. BMC Genomics. 2018. PMID: 29334892 Free PMC article. - The Expanding Family of Virophages.
Bekliz M, Colson P, La Scola B. Bekliz M, et al. Viruses. 2016 Nov 23;8(11):317. doi: 10.3390/v8110317. Viruses. 2016. PMID: 27886075 Free PMC article. Review. - Ecogenomics of virophages and their giant virus hosts assessed through time series metagenomics.
Roux S, Chan LK, Egan R, Malmstrom RR, McMahon KD, Sullivan MB. Roux S, et al. Nat Commun. 2017 Oct 11;8(1):858. doi: 10.1038/s41467-017-01086-2. Nat Commun. 2017. PMID: 29021524 Free PMC article. - Discovery and Further Studies on Giant Viruses at the IHU Mediterranee Infection That Modified the Perception of the Virosphere.
Rolland C, Andreani J, Louazani AC, Aherfi S, Francis R, Rodrigues R, Silva LS, Sahmi D, Mougari S, Chelkha N, Bekliz M, Silva L, Assis F, Dornas F, Khalil JYB, Pagnier I, Desnues C, Levasseur A, Colson P, Abrahão J, La Scola B. Rolland C, et al. Viruses. 2019 Mar 30;11(4):312. doi: 10.3390/v11040312. Viruses. 2019. PMID: 30935049 Free PMC article. Review.
References
- Adriaenssens E.M., Van Zyl L., De Maayer P., Rubagotti E., Rybicki E., Tuffin M., Cowan D.A. Metagenomic analysis of the viral community in Namib Desert hypoliths. Environ Microbiol. 2014;17:480–495. - PubMed
- Altschup S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Bamford D.H., Grimes J.M., Stuart D.I. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15:655–663. - PubMed
- Desnues C., Boyer M., Raoult D. Sputnik, a virophage infecting the viral domain of life. Adv Virus Res. 2012;82:63–89. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources