Squamous Cell Cancers: A Unified Perspective on Biology and Genetics - PubMed (original) (raw)
Review
Squamous Cell Cancers: A Unified Perspective on Biology and Genetics
G Paolo Dotto et al. Cancer Cell. 2016.
Abstract
Squamous cell carcinomas (SCCs) represent the most frequent human solid tumors and are a major cause of cancer mortality. These highly heterogeneous tumors arise from closely interconnected epithelial cell populations with intrinsic self-renewal potential inversely related to the stratified differentiation program. SCCs can also originate from simple or pseudo-stratified epithelia through activation of quiescent cells and/or a switch in cell-fate determination. Here, we focus on specific determinants implicated in the development of SCCs by recent large-scale genomic, genetic, and epigenetic studies, and complementary functional analysis. The evidence indicates that SCCs from various body sites, while clinically treated as separate entities, have common determinants, pointing to a unified perspective of the disease and potential new avenues for prevention and treatment.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
Figure 1. Incidence and mortality for major SCC types
Statistics calculated from: Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray, F. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. Lyon, France: International Agency for Research on Cancer; 2013. Available from:
, accessed on 27/09/2015.
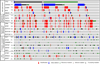
Figure 2. Pattern of frequently altered genes (>5% frequency) in HNSCC
The results shown here are based upon data generated by the cBioPortal for Cancer Genomics (
http://www.cbioportal.org/index.do
) (Cerami et al., 2012; Gao et al., 2013), and represent the genes relevant to this review with >5% alteration frequency. The dataset included all tumor samples with sequencing and CNA data (n=279). Mutually exclusive alterations were found for 94 gene pairs, only 2 of which statistically significant: TP53, EP300 (p = 0.01) and KMT2D – NFE2L2 (p = 0.038).
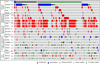
Figure 3. Pattern of frequently altered genes (>5% frequency) in LSCC
The results shown here are based upon data generated by the cBioPortal for Cancer Genomics (
http://www.cbioportal.org/index.do
) (Cerami et al., 2012; Gao et al., 2013), and represent the genes relevant to this review with >5% alteration frequency. The dataset included all tumor samples with sequencing and CNA data (n=178). Mutually exclusive alterations were found for 90 gene pairs, only 1 of which statistically significant: TP53 – KMT2C (p = 0.017).
Figure 4. Specific pattern and mapping of Notch1, Notch2 and Notch3 mutations in HNSCCs
The results shown here are based upon data generated by the cBioPortal for Cancer Genomics (
http://www.cbioportal.org/index.do
) (Cerami et al., 2012; Gao et al., 2013). Boxes of various colors correspond to different domains of the Notch proteins as evidenced on the cbioportal website according to the Pfam data base annotation (
http://pfam.xfam.org/family/PF06816
) as follows : EGF repeats (green); Lin-12/Notch repeats, LNR (yellow); heterodimerization domain, HD (purple); transmembrane region, TM (blue lines); ankyrin repeats, ANK (orange); PEST domain (light green).
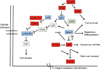
Figure 5. Model of potential interactions of epigenetic and genetic alterations in SCC development
Schematic of potential interactions amongst epigenetic and genetic alterations that may contribute, directly and indirectly, to SCC initiation and progression. Proteins with commonly accepted tumor promoting and suppressing functions are highlighted in red and blue respectively, while a protein involved in epigenetic regulation, p300, is highlighted in green.
References
- Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, Solari A, Bobisse S, Rondina MB, Guzzardo V, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. - PubMed
- Armstrong BK, Kricker A. The epidemiology of UV induced skin cancer. J Photochem Photobiol B. 2001;63:8–18. - PubMed
- Arnault JP, Mateus C, Escudier B, Tomasic G, Wechsler J, Hollville E, Soria JC, Malka D, Sarasin A, Larcher M, et al. Skin tumors induced by sorafenib; paradoxic RAS-RAF pathway activation and oncogenic mutations of HRAS, TP53, and TGFBR1. Clin Cancer Res. 2012;18:263–272. - PubMed
Publication types
MeSH terms
Grants and funding
- P30 DK050306/DK/NIDDK NIH HHS/United States
- R01 AR064786/AR/NIAMS NIH HHS/United States
- R01 AR039190/AR/NIAMS NIH HHS/United States
- P01 CA098101/CA/NCI NIH HHS/United States
- P30 CA016520/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases