Metabolic Model-Based Integration of Microbiome Taxonomic and Metabolomic Profiles Elucidates Mechanistic Links between Ecological and Metabolic Variation - PubMed (original) (raw)
Metabolic Model-Based Integration of Microbiome Taxonomic and Metabolomic Profiles Elucidates Mechanistic Links between Ecological and Metabolic Variation
Cecilia Noecker et al. mSystems. 2016 Jan-Feb.
Abstract
Multiple molecular assays now enable high-throughput profiling of the ecology, metabolic capacity, and activity of the human microbiome. However, to date, analyses of such multi-omic data typically focus on statistical associations, often ignoring extensive prior knowledge of the mechanisms linking these various facets of the microbiome. Here, we introduce a comprehensive framework to systematically link variation in metabolomic data with community composition by utilizing taxonomic, genomic, and metabolic information. Specifically, we integrate available and inferred genomic data, metabolic network modeling, and a method for predicting community-wide metabolite turnover to estimate the biosynthetic and degradation potential of a given community. Our framework then compares variation in predicted metabolic potential with variation in measured metabolites' abundances to evaluate whether community composition can explain observed shifts in the community metabolome, and to identify key taxa and genes contributing to the shifts. Focusing on two independent vaginal microbiome data sets, each pairing 16S community profiling with large-scale metabolomics, we demonstrate that our framework successfully recapitulates observed variation in 37% of metabolites. Well-predicted metabolite variation tends to result from disease-associated metabolism. We further identify several disease-enriched species that contribute significantly to these predictions. Interestingly, our analysis also detects metabolites for which the predicted variation negatively correlates with the measured variation, suggesting environmental control points of community metabolism. Applying this framework to gut microbiome data sets reveals similar trends, including prediction of bile acid metabolite shifts. This framework is an important first step toward a system-level multi-omic integration and an improved mechanistic understanding of the microbiome activity and dynamics in health and disease.
Importance: Studies characterizing both the taxonomic composition and metabolic profile of various microbial communities are becoming increasingly common, yet new computational methods are needed to integrate and interpret these data in terms of known biological mechanisms. Here, we introduce an analytical framework to link species composition and metabolite measurements, using a simple model to predict the effects of community ecology on metabolite concentrations and evaluating whether these predictions agree with measured metabolomic profiles. We find that a surprisingly large proportion of metabolite variation in the vaginal microbiome can be predicted based on species composition (including dramatic shifts associated with disease), identify putative mechanisms underlying these predictions, and evaluate the roles of individual bacterial species and genes. Analysis of gut microbiome data using this framework recovers similar community metabolic trends. This framework lays the foundation for model-based multi-omic integrative studies, ultimately improving our understanding of microbial community metabolism.
Keywords: community composition; metabolic modeling; metabolomics; microbiome; multi-omic.
Figures
FIG 1
Framework for integrating taxonomic and metabolomic data. Species composition is first used to predict the metagenome’s gene content, which is then paired with reaction information to estimate the community metabolic potential (CMP) for each sample and metabolite. Variation in predicted CMP scores is compared to variation in measured metabolite abundances (using pairwise differences) to identify well-predicted metabolites. A perturbation-based approach is used to additionally identify key species, gene, and reaction contributors to CMP scores.
FIG 2
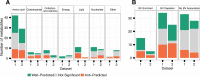
Metabolite predictability across metabolic categories (A) and disease states (B) in the vaginal microbiome. Well-predicted metabolites are defined as those for which variation in CMP scores is significantly correlated (using a Mantel test) with variation in measured metabolite abundance at a false discovery rate (FDR) of 0.01. Anti-predicted metabolites are similarly defined as those for which variation in CMP scores is significantly negatively correlated with variation in measured metabolite abundances (FDR 0.01). Metabolic categorization is based on KEGG data, and disease enrichment is based on a Wilcoxon rank sum test for association with bacterial vaginosis (BV) with a Bonferroni-corrected P value of <0.1.
FIG 3
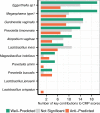
Key species contributors to metabolites in the vaginal microbiome. Each species that participated in the calculation of CMP scores in data set 1 is shown along the y axis. The x axis indicates the numbers of well-predicted and anti-predicted metabolites (as well as those with nonsignificant predictions) for which that species was a key contributor (see Materials and Methods).
FIG 4
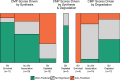
Trends in metabolite predictability in terms of key gene contributors. Area plots depict the numbers of metabolites in data set 1 whose CMP scores are driven by synthesis, by degradation, or by both in relation to their association with the host state and their predictability. The width of each box corresponds to the number of metabolites associated with each host disease state (enriched in BV samples, depleted in BV samples, or neither), and the height corresponds to the number of metabolites that are well-predicted, anti-predicted, or not significantly predicted (also indicated by color). See Fig. S6 in the supplemental material for a similar plot describing metabolite prediction in data set 2.
FIG 5
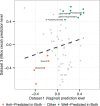
Metabolite predictability is consistent between vaginal and mouse cecal data sets. The plot shows the relationship between the level of predictability for each metabolite (measured as the Spearman correlation between pairwise differences in calculated CMP scores and pairwise differences in measured metabolite abundances) in data set 1 (human vaginal microbiome samples) and data set 3 (mouse gut samples). Colors indicate metabolites that are well-predicted in both data sets or anti-predicted in both data sets. Metabolites that are well-predicted in both data sets are enriched for amino acid catabolites, including phenylacetate, spermidine, and beta-alanine.
Similar articles
- A meta-analysis study of the robustness and universality of gut microbiome-metabolome associations.
Muller E, Algavi YM, Borenstein E. Muller E, et al. Microbiome. 2021 Oct 12;9(1):203. doi: 10.1186/s40168-021-01149-z. Microbiome. 2021. PMID: 34641974 Free PMC article. - Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.
Wolahan SM, Hirt D, Glenn TC. Wolahan SM, et al. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review. - Defining and Evaluating Microbial Contributions to Metabolite Variation in Microbiome-Metabolome Association Studies.
Noecker C, Chiu HC, McNally CP, Borenstein E. Noecker C, et al. mSystems. 2019 Dec 17;4(6):e00579-19. doi: 10.1128/mSystems.00579-19. mSystems. 2019. PMID: 31848305 Free PMC article. - MIMOSA2: a metabolic network-based tool for inferring mechanism-supported relationships in microbiome-metabolome data.
Noecker C, Eng A, Muller E, Borenstein E. Noecker C, et al. Bioinformatics. 2022 Mar 4;38(6):1615-1623. doi: 10.1093/bioinformatics/btac003. Bioinformatics. 2022. PMID: 34999748 Free PMC article. - Towards a predictive systems-level model of the human microbiome: progress, challenges, and opportunities.
Greenblum S, Chiu HC, Levy R, Carr R, Borenstein E. Greenblum S, et al. Curr Opin Biotechnol. 2013 Aug;24(4):810-20. doi: 10.1016/j.copbio.2013.04.001. Epub 2013 Apr 23. Curr Opin Biotechnol. 2013. PMID: 23623295 Free PMC article. Review.
Cited by
- Pathway-Based Integrative Analysis of Metabolome and Microbiome Data from Hepatocellular Carcinoma and Liver Cirrhosis Patients.
Kim B, Cho EJ, Yoon JH, Kim SS, Cheong JY, Cho SW, Park T. Kim B, et al. Cancers (Basel). 2020 Sep 21;12(9):2705. doi: 10.3390/cancers12092705. Cancers (Basel). 2020. PMID: 32967314 Free PMC article. - Gut Microbial Perturbation and Host Response Induce Redox Pathway Upregulation along the Gut-Liver Axis during Giardiasis in C57BL/6J Mouse Model.
Karpe AV, Hutton ML, Mileto SJ, James ML, Evans C, Ghodke AB, Shah RM, Metcalfe SS, Liu JW, Walsh T, Lyras D, Palombo EA, Beale DJ. Karpe AV, et al. Int J Mol Sci. 2023 Jan 13;24(2):1636. doi: 10.3390/ijms24021636. Int J Mol Sci. 2023. PMID: 36675151 Free PMC article. - The metaRbolomics Toolbox in Bioconductor and beyond.
Stanstrup J, Broeckling CD, Helmus R, Hoffmann N, Mathé E, Naake T, Nicolotti L, Peters K, Rainer J, Salek RM, Schulze T, Schymanski EL, Stravs MA, Thévenot EA, Treutler H, Weber RJM, Willighagen E, Witting M, Neumann S. Stanstrup J, et al. Metabolites. 2019 Sep 23;9(10):200. doi: 10.3390/metabo9100200. Metabolites. 2019. PMID: 31548506 Free PMC article. Review. - Progressive microbial adaptation of the bovine rumen and hindgut in response to a step-wise increase in dietary starch and the influence of phytogenic supplementation.
Ricci S, Pacífico C, Castillo-Lopez E, Rivera-Chacon R, Schwartz-Zimmermann HE, Reisinger N, Berthiller F, Zebeli Q, Petri RM. Ricci S, et al. Front Microbiol. 2022 Jul 22;13:920427. doi: 10.3389/fmicb.2022.920427. eCollection 2022. Front Microbiol. 2022. PMID: 35935232 Free PMC article. - Taxa-function robustness in microbial communities.
Eng A, Borenstein E. Eng A, et al. Microbiome. 2018 Mar 2;6(1):45. doi: 10.1186/s40168-018-0425-4. Microbiome. 2018. PMID: 29499759 Free PMC article.
References
- Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, Kim SG, Li H, Gao Z, Mahana D, Zárate Rodriguez JG, Rogers AB, Robine N, Loke P, Blaser MJ. 2014. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158:705–721. doi:10.1016/j.cell.2014.05.052. - DOI - PMC - PubMed
- Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. 2013. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 19:576–585. doi:10.1038/nm.3145. - DOI - PMC - PubMed
Grants and funding
- DP2 AT007802/AT/NCCIH NIH HHS/United States
- K01 GM109236/GM/NIGMS NIH HHS/United States
- R01 AI061628/AI/NIAID NIH HHS/United States
- R01 HG005816/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources