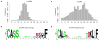Landscape of tumor-infiltrating T cell repertoire of human cancers - PubMed (original) (raw)
doi: 10.1038/ng.3581. Epub 2016 May 30.
Taiwen Li 1 3, Jean-Christophe Pignon 4, Binbin Wang 5, Jinzeng Wang 5, Sachet A Shukla 6, Ruoxu Dou 7, Qianming Chen 3, F Stephen Hodi 8, Toni K Choueiri 9, Catherine Wu 6, Nir Hacohen 10, Sabina Signoretti 4, Jun S Liu 2, X Shirley Liu 1 2
Affiliations
- PMID: 27240091
- PMCID: PMC5298896
- DOI: 10.1038/ng.3581
Landscape of tumor-infiltrating T cell repertoire of human cancers
Bo Li et al. Nat Genet. 2016 Jul.
Abstract
We developed a computational method to infer the complementarity-determining region 3 (CDR3) sequences of tumor-infiltrating T cells in 9,142 RNA-seq samples across 29 cancer types. We identified over 600,000 CDR3 sequences, including 15% that were full length. CDR3 sequence length distribution and amino acid conservation, as well as variable gene usage, for infiltrating T cells in many tumors, except in brain and kidney cancers, resembled those for peripheral blood cells from healthy donors. We observed a strong association between T cell diversity and tumor mutation load, and we predicted SPAG5 and TSSK6 as putative immunogenic cancer/testis antigens in multiple cancers. Finally, we identified three potential immunogenic somatic mutations on the basis of their co-occurrence with CDR3 sequences. One of them, a PRAMEF4 mutation encoding p.Phe300Val, was predicted to result in peptide binding strongly to both MHC class I and class II molecules, with matched HLA types in its carriers. Our analyses have the potential to simultaneously identify immunogenic neoantigens and tumor-reactive T cell clonotypes.
Conflict of interest statement
Competing financial interests: the authors declared no competing financial interest.
Figures
Figure 1
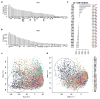
Distribution of αβ T cell variable gene usage and γδ T cell abundance in multiple cancer types. a–b: Proportions of TRAV and TRBV genes in decreasing order. IMGT functional genes were selected in the display. c–d. PCA analysis on TRAV and TRBV usage across different cancer types. For TRAV, PC1 was driven by the difference between brain cancer (LGG) and other tumors, while PC2 was driven by kidney cancer (KIRC). Dark blue circle: LGG samples; cyan circle: KIRC samples. e. γδ T cell fractions (labeled in the x-axis) in multiple cancer types in decreasing order. The mean γδ T cell fraction across all samples was 4.8%. For each cancer, we used Binomial test with expected probability 0.048 to calculate the statistical significance. We applied Benjamini-Hochberg adjusted P values for FDR. The numbers listed on the right margin of the plot are q values. Disease abbreviations: ACC: adenocortical carcinoma, BLCA: bladder carcinoma, BRCA: breast carcinoma, CESC: cervical squamous carcinoma, CHOL: cholangiocarcinoma, COAD: colon adenocarcinoma, DLBC: diffusive large B-cell lymphoma, GBM: glioblastoma multiforme, HNSC: head and neck carcinoma, KICH: kidney chromophobe, KIRC: kidney renal clear cell carcinoma, KIRP: kidney renal papillary cell carcinoma, LGG: lower grade glioma, LIHC: liver hepatocellular carcinoma, LUAD: lung adenocarcinoma, LUSC: lung squamous carcinoma, MESO: mesothelioma, PAAD: pancreatic adenocarcinoma, PCPG: pheochromocytoma and paraganglioma, PRAD: prostate adenocarcinoma, READ: rectum adenocarcinoma, SARC: sarcoma, SKCM: skin cutaneous melanoma, TGCT: testicular germ cell tumors, THCA: thyroid carcinoma, THYM: thymoma, UCEC: uterine corpus endometrial carcinoma, UCS: uterine carsinosarcoma, UVM: uveal melanoma.
Figure 2
Length and amino acid conservation of β and δ chain CDR3 sequences in tumor-infiltrating T cells. Length distribution of complete CDR3 calls was estimated using histogram for β and δ chains (a and c). Length 14 β-CDR3 and length 20 δ-CDR3 sequences were selected for weblogo analysis (b and d). The y-axis in the sequence logo plot was the conservation score. For a given locus, the height of a letter reflects the relative frequency of that amino acid.
Figure 3
Public and private β-CDR3 amino acid sequences have different lengths and hydrophobicity. a. β-CDR3 sharing between TCGA tumor, TCGA normal samples and peripheral blood repertoire displayed in Venn diagram. Colors indicated difference tissues: red: peripheral blood; green: TCGA normal samples; blue: TCGA tumor samples. The numbers of sharing labeled inside the eclipses were the overall calls of that category. Numbers labeled outside and connected to a colored region is the counts of the overlapped calls between categories. b. Distribution of public β-CDR3 frequency. Sequence sharing was determined using only TCGA data, not including the 10,249 sequences shared with blood repertoire. c. Comparison of β-CDR3 lengths of private and public sequences. P value was calculated using Wilcoxon test. Box includes data between the 25th and 75th percentiles, with horizontal line indicates the median. There are 14,443 and 51,583 sequences in the public and private group respectively. d. Hydrophobicity analysis of the middle 3 amino acids (see main text for details) in the private and public CDR3 sequences. In each position, Binomial test was applied to estimate the significance of the difference in hydrophobic amino acid fraction between groups, using the fraction in the public group as expected probability. All three positions were significant at FDR=0.05.
Figure 4
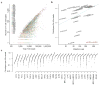
The diversity of T cell clonotypes positively associates with cancer somatic mutation load. a. Scatter plot of the number of CDR3 calls in each sample against the total reads extracted from the 3 TCR regions. Prostate and pancreatic cancers were excluded due to high expression of non-TCR genes in the region (Methods). b. Clonotypes per kilo-reads (CPK) was positively associated with tumor mutation load. Median CPK and median somatic mutation load for each cancer type were displayed on the scatter plot. Cancers with <50 samples were excluded. Significance was estimated using Spearman’s correlation test. c. Distributions of CPK across all cancer types. PAM50 subtypes of breast cancer46 were displayed to show the inter-tumor heterogeneity in this disease.
Figure 5
Association of T cell diversity and expression of cancer/testis antigens reveals SPAG5 and TSSK6 as vaccine targets. Gray entries indicated the gene was not overexpressed in the tumor cells, as suggested by correlative analysis with tumor purity. Association between CPK and the CT antigen expression was evaluated using partial Spearman correlation corrected for tumor purity. Solid boxes indicated significant associations at FDR=0.2.
Figure 6
Non-synonymous mutations co-occur with CDR3 motif. a. Three pairs of NS mutation and CDR3 motifs co-occurred more often than random, with statistical significance (FDR=0.05) based on permutation test (Methods). MHC-I binding predictions were performed on all the possible 9 amino acid peptides derived from the above three mutations using NetMHC4.0, and those with mutated peptides binding stronger than wild type peptides were displayed (b). Bold letters indicated mutated amino acids.
Similar articles
- Characterization of the T-cell receptor beta chain repertoire in tumor-infiltrating lymphocytes.
Nakanishi K, Kukita Y, Segawa H, Inoue N, Ohue M, Kato K. Nakanishi K, et al. Cancer Med. 2016 Sep;5(9):2513-21. doi: 10.1002/cam4.828. Epub 2016 Jul 27. Cancer Med. 2016. PMID: 27465739 Free PMC article. - Immunological ignorance is an enabling feature of the oligo-clonal T cell response to melanoma neoantigens.
Linette GP, Becker-Hapak M, Skidmore ZL, Baroja ML, Xu C, Hundal J, Spencer DH, Fu W, Cummins C, Robnett M, Kaabinejadian S, Hildebrand WH, Magrini V, Demeter R, Krupnick AS, Griffith OL, Griffith M, Mardis ER, Carreno BM. Linette GP, et al. Proc Natl Acad Sci U S A. 2019 Nov 19;116(47):23662-23670. doi: 10.1073/pnas.1906026116. Epub 2019 Nov 4. Proc Natl Acad Sci U S A. 2019. PMID: 31685621 Free PMC article. - In-vivo dominant immune responses in aplastic anaemia: molecular tracking of putatively pathogenetic T-cell clones by TCR beta-CDR3 sequencing.
Risitano AM, Maciejewski JP, Green S, Plasilova M, Zeng W, Young NS. Risitano AM, et al. Lancet. 2004 Jul 24-30;364(9431):355-64. doi: 10.1016/S0140-6736(04)16724-X. Lancet. 2004. PMID: 15276395 - TCR repertoires of intratumoral T-cell subsets.
Linnemann C, Mezzadra R, Schumacher TN. Linnemann C, et al. Immunol Rev. 2014 Jan;257(1):72-82. doi: 10.1111/imr.12140. Immunol Rev. 2014. PMID: 24329790 Review. - Applications of Immunogenomics to Cancer.
Liu XS, Mardis ER. Liu XS, et al. Cell. 2017 Feb 9;168(4):600-612. doi: 10.1016/j.cell.2017.01.014. Cell. 2017. PMID: 28187283 Free PMC article. Review.
Cited by
- Cancer systems immunology.
Reticker-Flynn NE, Engleman EG. Reticker-Flynn NE, et al. Elife. 2020 Jul 13;9:e53839. doi: 10.7554/eLife.53839. Elife. 2020. PMID: 32657757 Free PMC article. Review. - STARTRAC analyses of scRNAseq data from tumor models reveal T cell dynamics and therapeutic targets.
Bhatt D, Kang B, Sawant D, Zheng L, Perez K, Huang Z, Sekirov L, Wolak D, Huang JY, Liu X, DeVoss J, Manzanillo PS, Pierce N, Zhang Z, Symons A, Ouyang W. Bhatt D, et al. J Exp Med. 2021 Jun 7;218(6):e20201329. doi: 10.1084/jem.20201329. J Exp Med. 2021. PMID: 33900375 Free PMC article. - Genomic Characterization of Six Virus-Associated Cancers Identifies Changes in the Tumor Immune Microenvironment and Altered Genetic Programs.
Varn FS, Schaafsma E, Wang Y, Cheng C. Varn FS, et al. Cancer Res. 2018 Nov 15;78(22):6413-6423. doi: 10.1158/0008-5472.CAN-18-1342. Epub 2018 Sep 25. Cancer Res. 2018. PMID: 30254145 Free PMC article. - Neoantigen Dissimilarity to the Self-Proteome Predicts Immunogenicity and Response to Immune Checkpoint Blockade.
Richman LP, Vonderheide RH, Rech AJ. Richman LP, et al. Cell Syst. 2019 Oct 23;9(4):375-382.e4. doi: 10.1016/j.cels.2019.08.009. Epub 2019 Oct 9. Cell Syst. 2019. PMID: 31606370 Free PMC article. - Best practices for bioinformatic characterization of neoantigens for clinical utility.
Richters MM, Xia H, Campbell KM, Gillanders WE, Griffith OL, Griffith M. Richters MM, et al. Genome Med. 2019 Aug 28;11(1):56. doi: 10.1186/s13073-019-0666-2. Genome Med. 2019. PMID: 31462330 Free PMC article. Review.
References
- Alt FW, et al. VDJ recombination. Immunol Today. 1992;13:306–14. - PubMed
- Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials