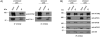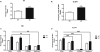Mammalian ataxin-2 modulates translation control at the pre-initiation complex via PI3K/mTOR and is induced by starvation - PubMed (original) (raw)
Mammalian ataxin-2 modulates translation control at the pre-initiation complex via PI3K/mTOR and is induced by starvation
Isabel Lastres-Becker et al. Biochim Biophys Acta. 2016 Sep.
Abstract
Ataxin-2 is a cytoplasmic protein, product of the ATXN2 gene, whose deficiency leads to obesity, while its gain-of-function leads to neural atrophy. Ataxin-2 affects RNA homeostasis, but its effects are unclear. Here, immunofluorescence analysis suggested that ataxin-2 associates with 48S pre-initiation components at stress granules in neurons and mouse embryonic fibroblasts, but is not essential for stress granule formation. Coimmunoprecipitation analysis showed associations of ataxin-2 with initiation factors, which were concentrated at monosome fractions of polysome gradients like ataxin-2, unlike its known interactor PABP. Mouse embryonic fibroblasts lacking ataxin-2 showed increased phosphorylation of translation modulators 4E-BP1 and ribosomal protein S6 through the PI3K-mTOR pathways. Indeed, human neuroblastoma cells after trophic deprivation showed a strong induction of ATXN2 transcript via mTOR inhibition. Our results support the notion that ataxin-2 is a nutritional stress-inducible modulator of mRNA translation at the pre-initiation complex.
Keywords: Amyotrophic lateral sclerosis; Diabetes mellitus; Spinocerebellar Ataxia type 2; TORC1; mRNA translation.
Copyright © 2016 Elsevier B.V. All rights reserved.
Figures
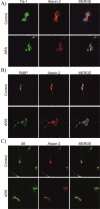
Figure 1. Ataxin-2 colocalizes with TIA-1, PABP, RPS6, eIF4G and eIF4A1 in SG of hippocampal neuron cultures
Hippocampal neurons after 7 days of culture (DIV7) were either left untreated (Control) or were stressed (ARS for arsenite, 0.5 mM for 45 min). The expression and subcellular localization of TIA-1 (A, F), PABP (B, F), RPS6 (C), eIF4G (D), eIF4A1 (E), and ataxin-2 (A-E) was monitored by immunofluorescence. Where indicated, the colocalization of two proteins was studied (Merge). DAPI staining was included to visualize the nuclei.
Figure 1. Ataxin-2 colocalizes with TIA-1, PABP, RPS6, eIF4G and eIF4A1 in SG of hippocampal neuron cultures
Hippocampal neurons after 7 days of culture (DIV7) were either left untreated (Control) or were stressed (ARS for arsenite, 0.5 mM for 45 min). The expression and subcellular localization of TIA-1 (A, F), PABP (B, F), RPS6 (C), eIF4G (D), eIF4A1 (E), and ataxin-2 (A-E) was monitored by immunofluorescence. Where indicated, the colocalization of two proteins was studied (Merge). DAPI staining was included to visualize the nuclei.
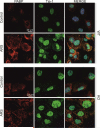
Figure 2. The absence of ataxin-2 in MEFs does not impair TIA-1 redistribution from nucleus to cytosolic SG
MEFs from WT and KO were either left untreated (Control) or were stressed (ARS for arsenite, 0.5 mM for 45 min). The subcellular localization of TIA-1 was monitored by immunofluorescence. Where indicated, the colocalization of two proteins was studied (Merge). DAPI staining was included to visualize the nuclei.
Figure 3. The absence of ataxin-2 does not prevent the formation of SG
MEFs from WT and KO were either left untreated (Control) or were stressed (ARS for arsenite, 0.5 mM for 45 min). The subcellular localization of PABP (A), RPS6 (B), eIF4G (C, E), eIF4A1 (D, E), and ataxin-2 (A-D) was monitored by immunofluorescence. Where indicated, the colocalization of two proteins was studied (Merge). DAPI staining was included to visualize the nuclei.
Figure 3. The absence of ataxin-2 does not prevent the formation of SG
MEFs from WT and KO were either left untreated (Control) or were stressed (ARS for arsenite, 0.5 mM for 45 min). The subcellular localization of PABP (A), RPS6 (B), eIF4G (C, E), eIF4A1 (D, E), and ataxin-2 (A-D) was monitored by immunofluorescence. Where indicated, the colocalization of two proteins was studied (Merge). DAPI staining was included to visualize the nuclei.
Figure 3. The absence of ataxin-2 does not prevent the formation of SG
MEFs from WT and KO were either left untreated (Control) or were stressed (ARS for arsenite, 0.5 mM for 45 min). The subcellular localization of PABP (A), RPS6 (B), eIF4G (C, E), eIF4A1 (D, E), and ataxin-2 (A-D) was monitored by immunofluorescence. Where indicated, the colocalization of two proteins was studied (Merge). DAPI staining was included to visualize the nuclei.
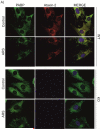
Figure 4. Ataxin-2 coimmunoprecipitates with TIA-1 and the translation pre-initiation complex
HEK-293 cells were left untreated or stressed with 0.5 mM arsenite for 45 min. Ataxin-2 was immunoprecipitated from cell homogenates with the 12SCA2-2B6 monoclonal antibody against ataxin-2 (IP ATXN2). Immunoprecipitates were analyzed by PAGE and western blotting with detection by the relevant antibodies. (A) Similar amounts of TIA-1 were coimmunoprecipitated with ataxin-2 in unstressed and stressed cells. (B) Similar amounts of several constituents of the translation pre-initiation complex were coimmunoprecipitated with ataxin-2 in unstressed and stressed cells.
Figure 5. Polysome gradients reveal ataxin-2 distribution and KO effects
(A, B) Polysome profiles on sucrose gradients: Lysates from WT-MEFs (A) and KO-MEFs (B) were layered on a 10-ml continuous sucrose gradient (10% to 50% sucrose). The profile of optical density representing the nucleotide content of the 40S, 60S, 80S ribosome components and polysomes are indicated. (C, D) Western blot analyses of the distribution of ataxin-2 and pre-initiation complex protein components among fractions from polysomal profiles in WT-MEFs (C) and KO-MEFs (D).
Figure 6. The absence of ataxin-2 induces increased phosphorylation of the ribosomal protein S6 and of the translation initiation repressor 4E-BP1, mainly through activation of the PI3K and the mTOR pathway
Phosphorylation of RPS6 (A) and 4E-BP1 (B) in WT- and KO-MEFs after 24 h of serum deprivation was measured using an ELISA kit, with the graphs illustrating the ratio of phospho-RPS6 versus total-RPS6 levels and of phospho-4E-BP1 versus total 4E-BP1. (C-D): Thirty min before cell harvest, MEF cultures were treated with the MEK inhibitor U0126, the PI3K inhibitor LY294002 or the mTOR inhibitor rapamycin; 5 WT- and 5 KO-MEF lines were tested in each case for RPS6 phosphorylation (C) and 4E-BP1 phosphorylation (D), with each experiment being performed three times (* p<0.05, ** p<0.01, *** p<0.005; * significant between WT-MEFs and KO-MEFs; # significant between KO-MEF different treatments). The relative results are presented with arbitrary units.
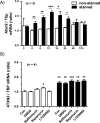
Figure 7. Nutrient and serum starvation elicits a transcriptional induction of ATXN2 via the mTOR pathway
(A) Human SH-SY5Y neuroblastoma cells were cultured in RPMI medium containing 10% FCS (RPMI + FCS) or in HBSS without nutrients (HBSS - FCS). At the indicated time points the ATXN2 transcript levels were quantified by qPCR and normalized to non-starvation (RPMI + FCS) at time 2 h. Student's t-test was used to calculate significance between starved and non-starved cells. (B) SH-SY5Y cells either treated with control (RPMI + FCS, white bars) or starvation medium (HBSS – FCS, black bars) with 0.05% DMSO, 5 nM bafilomycin A1 (BAF), 0.5 μM rapamycin as mTOR pathway antagonist (RAP), 25 μM LY294002 (LY) or nothing (CON) were incubated for 16 h prior to mRNA analysis by qPCR (* p<0.05, ** p<0.01, *** p<0.005).
Similar articles
- Genetic ablation of ataxin-2 increases several global translation factors in their transcript abundance but decreases translation rate.
Fittschen M, Lastres-Becker I, Halbach MV, Damrath E, Gispert S, Azizov M, Walter M, Müller S, Auburger G. Fittschen M, et al. Neurogenetics. 2015 Jul;16(3):181-92. doi: 10.1007/s10048-015-0441-5. Epub 2015 Feb 27. Neurogenetics. 2015. PMID: 25721894 Free PMC article. - Activation of the p70 S6 kinase and phosphorylation of the 4E-BP1 repressor of mRNA translation by type I interferons.
Lekmine F, Uddin S, Sassano A, Parmar S, Brachmann SM, Majchrzak B, Sonenberg N, Hay N, Fish EN, Platanias LC. Lekmine F, et al. J Biol Chem. 2003 Jul 25;278(30):27772-80. doi: 10.1074/jbc.M301364200. Epub 2003 May 20. J Biol Chem. 2003. PMID: 12759354 - Resistance exercise increases muscle protein synthesis and translation of eukaryotic initiation factor 2Bepsilon mRNA in a mammalian target of rapamycin-dependent manner.
Kubica N, Bolster DR, Farrell PA, Kimball SR, Jefferson LS. Kubica N, et al. J Biol Chem. 2005 Mar 4;280(9):7570-80. doi: 10.1074/jbc.M413732200. Epub 2004 Dec 10. J Biol Chem. 2005. PMID: 15591312 - Unraveling the Role of Ataxin-2 in Metabolism.
Carmo-Silva S, Nobrega C, Pereira de Almeida L, Cavadas C. Carmo-Silva S, et al. Trends Endocrinol Metab. 2017 Apr;28(4):309-318. doi: 10.1016/j.tem.2016.12.006. Epub 2017 Jan 21. Trends Endocrinol Metab. 2017. PMID: 28117213 Review. - Efficient Prevention of Neurodegenerative Diseases by Depletion of Starvation Response Factor Ataxin-2.
Auburger G, Sen NE, Meierhofer D, Başak AN, Gitler AD. Auburger G, et al. Trends Neurosci. 2017 Aug;40(8):507-516. doi: 10.1016/j.tins.2017.06.004. Epub 2017 Jul 3. Trends Neurosci. 2017. PMID: 28684172 Review.
Cited by
- Mouse Ataxin-2 Expansion Downregulates CamKII and Other Calcium Signaling Factors, Impairing Granule-Purkinje Neuron Synaptic Strength.
Arsović A, Halbach MV, Canet-Pons J, Esen-Sehir D, Döring C, Freudenberg F, Czechowska N, Seidel K, Baader SL, Gispert S, Sen NE, Auburger G. Arsović A, et al. Int J Mol Sci. 2020 Sep 12;21(18):6673. doi: 10.3390/ijms21186673. Int J Mol Sci. 2020. PMID: 32932600 Free PMC article. - The stress granule protein G3BP1 alleviates spinocerebellar ataxia-associated deficits.
Koppenol R, Conceição A, Afonso IT, Afonso-Reis R, Costa RG, Tomé S, Teixeira D, da Silva JP, Côdesso JM, Brito DVC, Mendonça L, Marcelo A, Pereira de Almeida L, Matos CA, Nóbrega C. Koppenol R, et al. Brain. 2023 Jun 1;146(6):2346-2363. doi: 10.1093/brain/awac473. Brain. 2023. PMID: 36511898 Free PMC article. - Ataxin-2: From RNA Control to Human Health and Disease.
Ostrowski LA, Hall AC, Mekhail K. Ostrowski LA, et al. Genes (Basel). 2017 Jun 5;8(6):157. doi: 10.3390/genes8060157. Genes (Basel). 2017. PMID: 28587229 Free PMC article. Review. - Control of CNS functions by RNA-binding proteins in neurological diseases.
Zhou Y, Dong F, Mao Y. Zhou Y, et al. Curr Pharmacol Rep. 2018 Aug;4(4):301-313. doi: 10.1007/s40495-018-0140-7. Epub 2018 May 2. Curr Pharmacol Rep. 2018. PMID: 30410853 Free PMC article. - Mechanisms contributing to adverse outcomes of COVID-19 in obesity.
Sudhakar M, Winfred SB, Meiyazhagan G, Venkatachalam DP. Sudhakar M, et al. Mol Cell Biochem. 2022 Apr;477(4):1155-1193. doi: 10.1007/s11010-022-04356-w. Epub 2022 Jan 27. Mol Cell Biochem. 2022. PMID: 35084674 Free PMC article. Review.
References
- Gispert S, Twells R, Orozco G, Brice A, Weber J, Heredero L, Scheufler K, Riley B, Allotey R, Nothers C, et al. Chromosomal assignment of the second locus for autosomal dominant cerebellar ataxia (SCA2) to chromosome 12q23-24.1. Nat Genet. 1993;4:295–299. - PubMed
- Imbert G, Saudou F, Yvert G, Devys D, Trottier Y, Garnier JM, Weber C, Mandel JL, Cancel G, Abbas N, Durr A, Didierjean O, Stevanin G, Agid Y, Brice A. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat Genet. 1996;14:285–291. - PubMed
- Pulst SM, Nechiporuk A, Nechiporuk T, Gispert S, Chen XN, Lopes-Cendes I, Pearlman S, Starkman S, Orozco-Diaz G, Lunkes A, DeJong P, Rouleau GA, Auburger G, Korenberg JR, Figueroa C, Sahba S. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat Genet. 1996;14:269–276. - PubMed
- Sanpei K, Takano H, Igarashi S, Sato T, Oyake M, Sasaki H, Wakisaka A, Tashiro K, Ishida Y, Ikeuchi T, Koide R, Saito M, Sato A, Tanaka T, Hanyu S, Takiyama Y, Nishizawa M, Shimizu N, Nomura Y, Segawa M, Iwabuchi K, Eguchi I, Tanaka H, Takahashi H, Tsuji S. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat Genet. 1996;14:277–284. - PubMed
- Estrada R, Galarraga J, Orozco G, Nodarse A, Auburger G. Spinocerebellar ataxia 2 (SCA2): morphometric analyses in 11 autopsies. Acta Neuropathol. 1999;97:306–310. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous