Herpes Simplex Virus Capsid Localization to ESCRT-VPS4 Complexes in the Presence and Absence of the Large Tegument Protein UL36p - PubMed (original) (raw)
Herpes Simplex Virus Capsid Localization to ESCRT-VPS4 Complexes in the Presence and Absence of the Large Tegument Protein UL36p
Himanshu Kharkwal et al. J Virol. 2016.
Abstract
UL36p (VP1/2) is the largest protein encoded by herpes simplex virus 1 (HSV-1) and resides in the innermost layer of tegument, the complex protein layer between the capsid and envelope. UL36p performs multiple functions in the HSV life cycle, including a critical but unknown role in capsid cytoplasmic envelopment. We tested whether UL36p is essential for envelopment because it is required to engage capsids with the cellular ESCRT/Vps4 apparatus. A green fluorescent protein (GFP)-fused form of the dominant negative ATPase Vps4-EQ was used to irreversibly tag ESCRT envelopment sites during infection by UL36p-expressing and UL36-null HSV strains. Using fluorescence microscopy and scanning electron microscopy, we quantitated capsid/Vps4-EQ colocalization and examined the ultrastructure of the corresponding viral assembly intermediates. We found that loss of UL36p resulted in a two-thirds reduction in the efficiency of capsid/Vps4-EQ association but that the remaining UL36p-null capsids were still able to engage the ESCRT envelopment apparatus. It appears that although UL36p helps to couple HSV capsids to the ESCRT pathway, this is likely not the sole reason for its absolute requirement for envelopment.
Importance: Envelopment of the HSV capsid is essential for the assembly of an infectious virion and requires the complex interplay of a large number of viral and cellular proteins. Critical to envelope assembly is the virally encoded protein UL36p, whose function is unknown. Here we test the hypothesis that UL36p is essential for the recruitment of cellular ESCRT complexes, which are also known to be required for envelopment.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
FIG 1
Preliminary characterization of UL36-null HSV strain GS3494. (A) PCR primers were used to amplify a 177-bp region of the bacterial chloramphenicol acetyltransferase (CAT) gene or a 403-bp region of the HSV UL19 gene, as indicated on the right. Lengths of DNA size markers (lane M), in base pairs, are shown on the left. Template DNA in each PCR was as follows: lane 1, total DNA from HS30 cells infected by UL36-null GS3494; lane 2, total DNA from HS30 cells infected by parental strain GS2822; lane 3, GS3494 BAC construct; lane 4, total DNA from uninfected HS30 cells. (B) Extracts of Vero cells infected by GS3494 (lanes 1) or the parental virus GS2822 (lanes 2) were subjected to SDS-PAGE and immunoblotted for UL36p, the major capsid protein VP5, and envelope proteins gB and gD, as indicated. (C) Vero (black bars) or HS30 (gray bars) cells were infected by GS2822 or GS3494 at an MOI of 0.01 and harvested after 72 h, and extracts were titrated on HS30 cell monolayers. Plaques formed by the UL36-null strain following replication on Vero cells likely correspond to residual input virus (propagated on HS30 cells).
FIG 2
A subset of HSV capsids are in close proximity to organellar membranes in intact cells in the absence of UL36p. Dishes of UL36p-expressing HS30 cells (A and B) or Vero cells (C to H) were infected by the UL36-null strain GS3494, fixed, and processed for thin-section electron microscopy. White arrows in panel D indicate examples of capsids on the edge of a large cytoplasmic cluster in close proximity to membranes. Black arrows indicate membrane-associated regions of electron density adjacent to capsids resembling tegument (E and G) or spokes (F and H) between the capsid and the lipid bilayer. Bars, 500 nm (C and D) and 200 nm (all other panels).
FIG 3
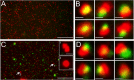
Representative fields of HSV capsid-associated and Vps4-EQ-GFP-bound cytoplasmic particles. HEK293 cells were induced via tetracycline to express Vps4-EQ-GFP and then infected with HSV for 16 h, a PNS was prepared, and virions and organelles were attached to MatTek dishes. After fixation, fields of particles were imaged in the red (VP26-mRFP1) and green (Vps4-EQ-GFP) channels; merged images are shown. (A) Low-magnification field of a PNS from GS2822-infected cells. (B) Representative gallery of six individual particles from a field similar to that in panel A. (C) Low-magnification field of a PNS from GS3494-infected cells. Two large red fluorescent structures (indicated by white arrows and numbered 1 and 2) are shown as higher-magnification insets. (D) Representative gallery of six individual particles from a field similar to that in panel C. Bars, 30 μm (A and C), 1 μm (insets in panel C), and 0.5 μm (B and D).
FIG 4
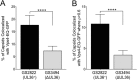
Effect of UL36p loss on capsid/Vps4-EQ colocalization. Vps4-EQ-GFP-expressing cells were infected with HSV GS2822 (UL36+; black bars) or GS3494 (ΔUL36; gray bars). Postnuclear supernatants (PNS) were prepared, attached to MatTek dishes, fixed, and imaged. (A) Percentages of all mRFP1-fluorescing particles that colocalized with Vps4-EQ-GFP. (B) Percentages of all mRFP1-fluorescing particles that exhibited colocalization with Vps4-EQ-GFP with ρ values of >0.6. For both panels A and B, plotted values show means and standard deviations. Data were analyzed by the Student t test (****, P < 0.0001).
FIG 5
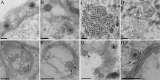
Correlative light and electron microscopic analysis of HSV capsid/Vps4-EQ-GFP colocalization in the presence and absence of UL36p. Cytoplasmic particles from GS2822-infected (A to F) or GS3494-infected (G to P) Vps4-EQ-GFP-expressing HEK293 cells were prepared and imaged as described in the legend to Fig. 3. Samples were then processed for scanning electron microscopy (SEM) and images aligned. Images are shown in pairs; for each pair, the left panel shows an alignment of fluorescent red (capsid) and green (Vps4-EQ-GFP) images with the SEM image, and the right panel shows the SEM image alone, usually at a higher resolution. White boxes in panels E and K correspond to a region of the structure shown at a higher resolution in panels F and L, respectively. Bars for SEM/fluorescence-aligned images, 100 nm (A and O) and 200 nm (C, E, G, I, K, and M); bars for SEM-only images (B, D, F, H, J, L, N, and P), 100 nm. Arrows in panels F, J, and L label capsids docked to organelles but apparently not engaged with the ESCRT apparatus. Arrowheads (B and N) indicate bead-like structures around the leading edge of the membrane.
Similar articles
- An ESCRT/VPS4 Envelopment Trap To Examine the Mechanism of Alphaherpesvirus Assembly and Transport in Neurons.
Barnes J, Jordan BA, Wilson DW. Barnes J, et al. J Virol. 2022 Mar 23;96(6):e0217821. doi: 10.1128/jvi.02178-21. Epub 2022 Jan 19. J Virol. 2022. PMID: 35045266 Free PMC article. - Seeking Closure: How Do Herpesviruses Recruit the Cellular ESCRT Apparatus?
Barnes J, Wilson DW. Barnes J, et al. J Virol. 2019 Jun 14;93(13):e00392-19. doi: 10.1128/JVI.00392-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30996099 Free PMC article. Review. - Herpes Simplex Virus Capsid-Organelle Association in the Absence of the Large Tegument Protein UL36p.
Kharkwal H, Furgiuele SS, Smith CG, Wilson DW. Kharkwal H, et al. J Virol. 2015 Nov;89(22):11372-82. doi: 10.1128/JVI.01893-15. Epub 2015 Sep 2. J Virol. 2015. PMID: 26339048 Free PMC article. - Conserved Tryptophan Motifs in the Large Tegument Protein pUL36 Are Required for Efficient Secondary Envelopment of Herpes Simplex Virus Capsids.
Ivanova L, Buch A, Döhner K, Pohlmann A, Binz A, Prank U, Sandbaumhüter M, Bauerfeind R, Sodeik B. Ivanova L, et al. J Virol. 2016 May 12;90(11):5368-5383. doi: 10.1128/JVI.03167-15. Print 2016 Jun 1. J Virol. 2016. PMID: 27009950 Free PMC article. - HSV-1 Cytoplasmic Envelopment and Egress.
Ahmad I, Wilson DW. Ahmad I, et al. Int J Mol Sci. 2020 Aug 19;21(17):5969. doi: 10.3390/ijms21175969. Int J Mol Sci. 2020. PMID: 32825127 Free PMC article. Review.
Cited by
- Motor Skills: Recruitment of Kinesins, Myosins and Dynein during Assembly and Egress of Alphaherpesviruses.
Wilson DW. Wilson DW. Viruses. 2021 Aug 17;13(8):1622. doi: 10.3390/v13081622. Viruses. 2021. PMID: 34452486 Free PMC article. Review. - Nuclear Egress.
Draganova EB, Thorsen MK, Heldwein EE. Draganova EB, et al. Curr Issues Mol Biol. 2021;41:125-170. doi: 10.21775/cimb.041.125. Epub 2020 Aug 7. Curr Issues Mol Biol. 2021. PMID: 32764158 Free PMC article. Review. - An ESCRT/VPS4 Envelopment Trap To Examine the Mechanism of Alphaherpesvirus Assembly and Transport in Neurons.
Barnes J, Jordan BA, Wilson DW. Barnes J, et al. J Virol. 2022 Mar 23;96(6):e0217821. doi: 10.1128/jvi.02178-21. Epub 2022 Jan 19. J Virol. 2022. PMID: 35045266 Free PMC article. - Global Proteomic Changes Induced by the Epstein-Barr Virus Oncoproteins Latent Membrane Protein 1 and 2A.
DeKroon RM, Gunawardena HP, Edwards R, Raab-Traub N. DeKroon RM, et al. mBio. 2018 Jun 19;9(3):e00959-18. doi: 10.1128/mBio.00959-18. mBio. 2018. PMID: 29921667 Free PMC article. - Seeking Closure: How Do Herpesviruses Recruit the Cellular ESCRT Apparatus?
Barnes J, Wilson DW. Barnes J, et al. J Virol. 2019 Jun 14;93(13):e00392-19. doi: 10.1128/JVI.00392-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30996099 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical