pong: fast analysis and visualization of latent clusters in population genetic data - PubMed (original) (raw)
pong: fast analysis and visualization of latent clusters in population genetic data
Aaron A Behr et al. Bioinformatics. 2016.
Abstract
Motivation: A series of methods in population genetics use multilocus genotype data to assign individuals membership in latent clusters. These methods belong to a broad class of mixed-membership models, such as latent Dirichlet allocation used to analyze text corpora. Inference from mixed-membership models can produce different output matrices when repeatedly applied to the same inputs, and the number of latent clusters is a parameter that is often varied in the analysis pipeline. For these reasons, quantifying, visualizing, and annotating the output from mixed-membership models are bottlenecks for investigators across multiple disciplines from ecology to text data mining.
Results: We introduce pong, a network-graphical approach for analyzing and visualizing membership in latent clusters with a native interactive D3.js visualization. pong leverages efficient algorithms for solving the Assignment Problem to dramatically reduce runtime while increasing accuracy compared with other methods that process output from mixed-membership models. We apply pong to 225 705 unlinked genome-wide single-nucleotide variants from 2426 unrelated individuals in the 1000 Genomes Project, and identify previously overlooked aspects of global human population structure. We show that pong outpaces current solutions by more than an order of magnitude in runtime while providing a customizable and interactive visualization of population structure that is more accurate than those produced by current tools.
Availability and implementation: pong is freely available and can be installed using the Python package management system pip. pong's source code is available at https://github.com/abehr/pong
Contact: aaron_behr@alumni.brown.edu or sramachandran@brown.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2016. Published by Oxford University Press.
Figures
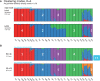
Fig. 1.
pong’s front end produces a D3.js visualization of maximum-weight alignments between runs, shown here for 20 Q matrices produced from clustering inference with ADMIXTURE (Alexander et al., 2009) applied to 1000 Genomes data (phase 3; Consortium, 2015). Each individual’s genome-wide ancestry within a barplot is depicted by K stacked colored lines. The left-to-right order of individuals is the same in each barplot. The barplots here are annotated with numbers (white) indicating which column of the underlying Q matrix is represented by a given cluster. (A) Characterizing modes at K = 4 by displaying the representative run of the major mode (here, k4r4) and the representative run of each minor mode. Three-letter population codes are shown at the bottom. (B) The maximum-weight alignment for the representative run for the major mode at K = 4 (k4r4, panel A) to that at K = 5. Membership in cluster 4 at K = 4 represents shared ancestry in East Asian and admixed American populations, and has been partitioned into Clusters 3 and 5 (representing East Asian and Native American ancestry, respectively) in the representative run of the major mode at K = 5
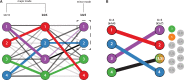
Fig. 2.
pong’s back-end model for the alignment of Q matrices, shown here from clustering inference with ADMIXTURE (Alexander et al., 2009) applied to 1000 Genomes data (phase 3; Consortium, 2015). Panel labels correspond to panels in Figure 1, and numbers in graph vertices correspond to the clusters labeled in Figure 1. (A) Characterizing modes from three runs of clustering inference at K = 4, the smallest K value with multiple modes for this dataset. Edge thickness corresponds to the value of pong’s default cluster similarity metric J (derived from Jaccard’s index; see
Supplementary Materials
), while edge opacity ranks connections for a cluster in run k4r4 to a cluster in run k4r3 (or in run k4r10). Note that both cluster 2 and 3 in k4r4 are most similar based on metric J to cluster 2 in k4r3; in order to find the maximum-weight perfect matching between the runs, pong matches cluster 3 in k4r4 with cluster 1 in k4r3. Bold labels indicate representative runs for the two modes. Seven other runs (not displayed for ease of visualization) are grouped in the same mode as k4r4 and k4r10; these nine runs comprise the major mode at K = 4 (Fig. 1A). k4r3 is the only run in the minor mode (Fig. 1A). (B) Alignment of representative runs for the major modes at K = 4 to K = 5. (52)=10 alignments are constructed between k4r4 and k5r7 (the representative run of the major mode at K = 5), constrained by the use of exactly one union node at K = 5. Of these 10 alignments, the alignment with maximum edge weight is shown and matches cluster 4 in k4r4 to the sum of clusters 3 and 5 in k5r7. The best matching for all other clusters are shown and informs the coloring of pong’s visualization (see Fig. 1B)
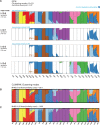
Fig. 3.
Visualizations of modes in population structure identified by pong and C
lumpak
at K = 10 for clustering inference with ADMIXTURE (Alexander et al., 2009) applied to 1000 Genomes data (phase 3; Consortium, 2015). The new cluster of membership coefficients first identified at K = 10 is denoted by light blue in each barplot. (A) pong’s dialog box of modes at K = 10, with multimodality highlighted. (B) C
lumpak
’s major mode at K = 10 averages over six runs of clustering inference output; the reported mean similarity score among these six runs is 0.811. South Asian (GIH), Han Chinese (CHB and CHS), and Puerto Rican (PUR) individuals all have ancestry depicted by the light blue cluster in this plot. The six runs averaged here are instead partitioned into three minor modes by pong in panel A. (C) C
lumpak
’s minor mode at K = 10 averages over four identical runs (mean similarity score is 1.000). This barplot contains the same information as the barplot of k4r10, representing pong’s major mode in panel A
Similar articles
- HitWalker2: visual analytics for precision medicine and beyond.
Bottomly D, McWeeney SK, Wilmot B. Bottomly D, et al. Bioinformatics. 2016 Apr 15;32(8):1253-5. doi: 10.1093/bioinformatics/btv739. Epub 2015 Dec 26. Bioinformatics. 2016. PMID: 26708334 Free PMC article. - Evaluating individual genome similarity with a topic model.
Juan L, Wang Y, Jiang J, Yang Q, Wang G, Wang Y. Juan L, et al. Bioinformatics. 2020 Sep 15;36(18):4757-4764. doi: 10.1093/bioinformatics/btaa583. Bioinformatics. 2020. PMID: 32573702 - Clumppling: cluster matching and permutation program with integer linear programming.
Liu X, Kopelman NM, Rosenberg NA. Liu X, et al. Bioinformatics. 2024 Jan 2;40(1):btad751. doi: 10.1093/bioinformatics/btad751. Bioinformatics. 2024. PMID: 38096585 Free PMC article. - PERF: an exhaustive algorithm for ultra-fast and efficient identification of microsatellites from large DNA sequences.
Avvaru AK, Sowpati DT, Mishra RK. Avvaru AK, et al. Bioinformatics. 2018 Mar 15;34(6):943-948. doi: 10.1093/bioinformatics/btx721. Bioinformatics. 2018. PMID: 29121165 - Granular clustering of de novo protein models.
Guzenko D, Strelkov SV. Guzenko D, et al. Bioinformatics. 2017 Feb 1;33(3):390-396. doi: 10.1093/bioinformatics/btw628. Bioinformatics. 2017. PMID: 28171609
Cited by
- Genomic diversity of the Japanese wheat core collection and selection of alleles for agronomic traits in the breeding process.
Mizuno N, Kobayashi F, Morita T, Handa H. Mizuno N, et al. Breed Sci. 2024 Jun;74(3):259-273. doi: 10.1270/jsbbs.23064. Epub 2024 Jun 25. Breed Sci. 2024. PMID: 39555015 Free PMC article. - Improving GWAS performance in underrepresented groups by appropriate modeling of genetics, environment, and sociocultural factors.
Cataldo-Ramirez CC, Lin M, Mcmahon A, Gignoux CR, Weaver TD, Henn BM. Cataldo-Ramirez CC, et al. bioRxiv [Preprint]. 2024 Oct 29:2024.10.28.620716. doi: 10.1101/2024.10.28.620716. bioRxiv. 2024. PMID: 39553939 Free PMC article. Preprint. - Exploring genetic diversity, population structure, and subgenome differences in the allopolyploid Camelina sativa: implications for future breeding and research studies.
Brock JR, Bird KA, Platts AE, Gomez-Cano F, Gupta SK, Palos K, Railey CE, Teresi SJ, Lee YS, Magallanes-Lundback M, Pawlowski EG, Nelson ADL, Grotewold E, Edger PP. Brock JR, et al. Hortic Res. 2024 Sep 9;11(11):uhae247. doi: 10.1093/hr/uhae247. eCollection 2024 Nov. Hortic Res. 2024. PMID: 39539416 Free PMC article. - Genetic diversity, population structure, and phylogeny of insular Spanish pepper landraces (Capsicum annuum L.) through phenotyping and genotyping-by-sequencing.
Ortega-Albero N, Barchi L, Fita A, Díaz M, Martínez F, Luna-Prohens JM, Rodríguez-Burruezo A. Ortega-Albero N, et al. Front Plant Sci. 2024 Oct 30;15:1435427. doi: 10.3389/fpls.2024.1435427. eCollection 2024. Front Plant Sci. 2024. PMID: 39539294 Free PMC article. - Nested admixture during and after the Trans-Atlantic Slave Trade on the island of São Tomé.
Ciccarella M, Laurent R, Szpiech ZA, Patin E, Dessarps-Freichey F, Utgé J, Lémée L, Semo A, Rocha J, Verdu P. Ciccarella M, et al. bioRxiv [Preprint]. 2024 Oct 23:2024.10.21.619344. doi: 10.1101/2024.10.21.619344. bioRxiv. 2024. PMID: 39484499 Free PMC article. Preprint.
References
- Blei D.M. et al. (2003) Latent Dirichlet Allocation. J. Mach. Learn. Res., 3, 993–1022.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources