PrimPol-A new polymerase on the block - PubMed (original) (raw)
Review
PrimPol-A new polymerase on the block
Sean G Rudd et al. Mol Cell Oncol. 2014.
Abstract
The DNA-directed primase-polymerase PrimPol of the archaeo-eukaryotic primase superfamily represents an ancient solution to the many problems faced during genome duplication. This versatile enzyme is capable of initiating de novo DNA/RNA synthesis, DNA chain elongation, and has the capacity to bypass modifications that stall the replisome by trans-lesion synthesis or origin-independent re-priming, thus allowing discontinuous synthesis of the leading strand. Recent studies have shown that PrimPol is an important new player in replication fork progression in eukaryotic cells; this review summarizes our current understanding of PrimPol and highlights important questions that remain to be addressed.
Keywords: AEP; DNA; PrimPol; TLS; lesions; polymerase; primase; replication; repriming; restart.
Figures
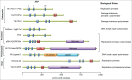
Figure 1.
The domain organization of members of the archaeo-eukaryotic primase superfamily. Domain organization of various AEPs is depicted: motifs I, II, and III of the catalytic AEP domain (blue boxes), zinc finger (Zn) motifs (red boxes), and additional domains often associated with AEP enzymes are shown. The putative or characterized role of each AEP is indicated. Domain organizations were deduced from the article by Iyer et al.
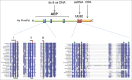
Figure 2.
Conserved domains and motifs present in the PrimPol family. The catalytic archaeo-eukaryotic primase (AEP) domain containing 3 signature motifs (I, II, and III; blue boxes), the UL52-like zinc finger domain (Zn), and the replication protein A (RPA)-interaction site (orange box) are indicated for human PrimPol, including amino acid numbers. Multiple sequence alignment was generated for a selection of PrimPol homologues; blue shading indicates ≥40% sequence identity, red circles indicate residues required for metal ion binding, orange circles indicate those required for nucleotide binding, and green circles those required for chelation of zinc.
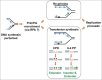
Figure 3.
PrimPol is a versatile DNA/RNA primase-polymerase in vitro. The reported in vitro activities of human PrimPol are represented schematically. Some activities are dependent on specific metal co-factors: for example, template scrunching to facilitate bypass and template-independent terminal transferase activity are only observed when manganese ions are used as co-factors. TLS, translesion synthesis; 8-oxo-guanine, 8-oxo-G; pyrimidine (6-4) pyrimidone photoproduct, 6-4 PP; apurinic/apyrimidinic site, AP site; cyclobutane pyrimidine dimer, CPD.
Figure 4.
Model of PrimPol-mediated replication fork progression. Distortion of DNA and base modification can be induced by various environmental insults and endogenous processes and, if not corrected prior to replication, can disrupt DNA synthesis by the cellular replicases (blue lines). A DNA modification on the leading strand is shown, which in this example has caused uncoupling of leading and lagging strand synthesis. This generates stretches of single-stranded DNA that will be coated by replication protein A (RPA), which in turn recruits PrimPol. PrimPol-dependent DNA or RNA synthesis (green lines) then facilitates restart of DNA replication. PrimPol may re-prime DNA synthesis downstream of the lesion leaving a daughter strand gap that can be subsequently filled by translesion synthesis (TLS) or homologous recombination (HR)-mediated processes. Alternatively, in the case of DNA lesions such as UV photoproducts (depicted in red lettering), PrimPol can use its TLS activity and directly extend the stalled primer terminus to synthesize DNA opposite the lesion, either alone or by cooperating with another DNA polymerase. For example, in the case of a template cyclobutane pyrimidine dimer (CPD), after incorporation of 2 terminal dA residues opposite the lesion, PrimPol can catalyze the extension of this mismatched terminus. In the case of a pyrimidine (6–4) pyrimidone photoproduct (6–4 PP), PrimPol can catalyze both the insertion of nucleotides opposite the damaged bases and the subsequent extension from the mismatched terminus, and thus could possibly catalyze complete bypass of this lesion. PrimPol misincorporates a dT opposite the 5′T of the lesion and either dG or dC opposite the 3′T, as shown in green.
Similar articles
- PrimPol-Prime Time to Reprime.
Guilliam TA, Doherty AJ. Guilliam TA, et al. Genes (Basel). 2017 Jan 6;8(1):20. doi: 10.3390/genes8010020. Genes (Basel). 2017. PMID: 28067825 Free PMC article. Review. - Repriming by PrimPol is critical for DNA replication restart downstream of lesions and chain-terminating nucleosides.
Kobayashi K, Guilliam TA, Tsuda M, Yamamoto J, Bailey LJ, Iwai S, Takeda S, Doherty AJ, Hirota K. Kobayashi K, et al. Cell Cycle. 2016 Aug 2;15(15):1997-2008. doi: 10.1080/15384101.2016.1191711. Epub 2016 May 26. Cell Cycle. 2016. PMID: 27230014 Free PMC article. - Human PrimPol activity is enhanced by RPA.
Martínez-Jiménez MI, Lahera A, Blanco L. Martínez-Jiménez MI, et al. Sci Rep. 2017 Apr 10;7(1):783. doi: 10.1038/s41598-017-00958-3. Sci Rep. 2017. PMID: 28396594 Free PMC article. - In vitro lesion bypass by human PrimPol.
Makarova AV, Boldinova EO, Belousova EA, Lavrik OI. Makarova AV, et al. DNA Repair (Amst). 2018 Oct;70:18-24. doi: 10.1016/j.dnarep.2018.07.009. Epub 2018 Jul 27. DNA Repair (Amst). 2018. PMID: 30098578 - Repriming DNA synthesis: an intrinsic restart pathway that maintains efficient genome replication.
Bainbridge LJ, Teague R, Doherty AJ. Bainbridge LJ, et al. Nucleic Acids Res. 2021 May 21;49(9):4831-4847. doi: 10.1093/nar/gkab176. Nucleic Acids Res. 2021. PMID: 33744934 Free PMC article. Review.
Cited by
- Translesion Synthesis: Insights into the Selection and Switching of DNA Polymerases.
Zhao L, Washington MT. Zhao L, et al. Genes (Basel). 2017 Jan 10;8(1):24. doi: 10.3390/genes8010024. Genes (Basel). 2017. PMID: 28075396 Free PMC article. Review. - Human PrimPol is a highly error-prone polymerase regulated by single-stranded DNA binding proteins.
Guilliam TA, Jozwiakowski SK, Ehlinger A, Barnes RP, Rudd SG, Bailey LJ, Skehel JM, Eckert KA, Chazin WJ, Doherty AJ. Guilliam TA, et al. Nucleic Acids Res. 2015 Jan;43(2):1056-68. doi: 10.1093/nar/gku1321. Epub 2014 Dec 29. Nucleic Acids Res. 2015. PMID: 25550423 Free PMC article. - REV1 coordinates a multi-faceted tolerance response to DNA alkylation damage and prevents chromosome shattering in Drosophila melanogaster.
Khodaverdian V, Sano T, Maggs LR, Tomarchio G, Dias A, Tran M, Clairmont C, McVey M. Khodaverdian V, et al. PLoS Genet. 2024 Jul 29;20(7):e1011181. doi: 10.1371/journal.pgen.1011181. eCollection 2024 Jul. PLoS Genet. 2024. PMID: 39074150 Free PMC article. - Translesion and Repair DNA Polymerases: Diverse Structure and Mechanism.
Yang W, Gao Y. Yang W, et al. Annu Rev Biochem. 2018 Jun 20;87:239-261. doi: 10.1146/annurev-biochem-062917-012405. Epub 2018 Mar 1. Annu Rev Biochem. 2018. PMID: 29494238 Free PMC article. Review. - The Intra-S Checkpoint Responses to DNA Damage.
Iyer DR, Rhind N. Iyer DR, et al. Genes (Basel). 2017 Feb 17;8(2):74. doi: 10.3390/genes8020074. Genes (Basel). 2017. PMID: 28218681 Free PMC article. Review.
References
- Aguilera A, Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 2008; 9:204-17; PMID:18227811; http://dx.doi.org/10.1038/nrg2268 - DOI - PubMed
- Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer 2008; 8:193-204; PMID:18256616; http://dx.doi.org/10.1038/nrc2342 - DOI - PubMed
- Wan L, Lou J, Xia Y, Su B, Liu T, Cui J, Sun Y, Lou H, Huang J. hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity. EMBO Rep 2013; 14:1104-12; PMID:24126761; http://dx.doi.org/10.1038/embor.2013.159 - DOI - PMC - PubMed
- García-Gómez S, Reyes A, Martínez-Jiménez MI, Chocrón ES, Mourón S, Terrados G, Powell C, Salido E, Méndez J, Holt IJ, et al. . PrimPol, an archaic primase/polymerase operating in human cells. Mol Cell 2013; 52:541-553; PMID:24207056; http://dx.doi.org/10.1016/j.molcel.2013.09.025 - DOI - PMC - PubMed
- Bianchi J, Rudd SG, Jozwiakowski SK, Bailey LJ, Soura V, Taylor E, Stevanovic I, Green AJ, Stracker TH, Lindsay HD, et al. . PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol Cell 2013; 52:566-73; PMID:24267451; http://dx.doi.org/10.1016/j.molcel.2013.10.035 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources