Predicting Ovarian Cancer Patients' Clinical Response to Platinum-Based Chemotherapy by Their Tumor Proteomic Signatures - PubMed (original) (raw)
Predicting Ovarian Cancer Patients' Clinical Response to Platinum-Based Chemotherapy by Their Tumor Proteomic Signatures
Kun-Hsing Yu et al. J Proteome Res. 2016.
Abstract
Ovarian cancer is the deadliest gynecologic malignancy in the United States with most patients diagnosed in the advanced stage of the disease. Platinum-based antineoplastic therapeutics is indispensable to treating advanced ovarian serous carcinoma. However, patients have heterogeneous responses to platinum drugs, and it is difficult to predict these interindividual differences before administering medication. In this study, we investigated the tumor proteomic profiles and clinical characteristics of 130 ovarian serous carcinoma patients analyzed by the Clinical Proteomic Tumor Analysis Consortium (CPTAC), predicted the platinum drug response using supervised machine learning methods, and evaluated our prediction models through leave-one-out cross-validation. Our data-driven feature selection approach indicated that tumor proteomics profiles contain information for predicting binarized platinum response (P < 0.0001). We further built a least absolute shrinkage and selection operator (LASSO)-Cox proportional hazards model that stratified patients into early relapse and late relapse groups (P = 0.00013). The top proteomic features indicative of platinum response were involved in ATP synthesis pathways and Ran GTPase binding. Overall, we demonstrated that proteomic profiles of ovarian serous carcinoma patients predicted platinum drug responses as well as provided insights into the biological processes influencing the efficacy of platinum-based therapeutics. Our analytical approach is also extensible to predicting response to other antineoplastic agents or treatment modalities for both ovarian and other cancers.
Keywords: bioinformatics; cancer biomarkers; drug resistance; ovarian cancer; tandem mass spectrometry.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Figure 1.
Protein expression levels were associated with platinum response of ovarian serous carcinoma patients. (A) Patients with similar platinum responses shared certain proteomic signatures. In this heatmap, each row is a patient, and each column is a protein. Hierarchical clustering results showed that platinum-resistant patients formed clusters, indicating that these protein signatures are correlated with platinum status of ovarian serous carcinoma patients. (B) Log expression levels of proteins associated with platinum response status. Abundance levels of 24 proteins, including KRT19, KRT4, ACTN4, RANBP1, IGLL5, and TPMT, were strongly associated with the clinical platinum response in ovarian serous carcinoma patients.
Figure 2.
Protein-protein interaction network revealed that proteins indicative of platinum response were significantly enriched in Ran-GTPase binding, ATP synthesis pathways, and regulation mechanisms of the cell cycle. The top proteins associated with platinum response were shown in color, and their interacting proteins were shown in gray.
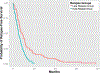
Figure 3.
Receiver operating characteristic (ROC) curve for platinum response prediction. Integrating the abundance levels of the top protein features identified in the training set, random forests, support vector machines, bagging, and naïve Bayes classifiers weakly predicted patients with different platinum responses on leave-one-out cross-validation with areas under the curves of approximately 0.58—0.64. These classifiers are significantly better than a null classifier (P = 7.96 × 10−9 for random forest with conditional inference trees, P = 2.10 × 10−7 for SVM with Gaussian kernel, P = 8.01 × 10−9 for SVM with polynomial kernel, P = 4.68 × 10−10 for naïve Bayes classifiers, P = 7.99 × 10−9 for Breiman’s random forest, and P = 7.99 × 10−9 for bagging). The black line indicates the performance of the null classifier. CIT: conditional inference trees.
Figure 4.
Proteomic signatures informed the platinum-free interval of patients. A LASSO-Cox proportional hazards model stratified ovarian serous carcinoma patients in the test set into two predicted relapse groups with a statistically significant difference in their platinum free interval (P = 0.00013 ± 0.00012).
Similar articles
- Incorporating SULF1 polymorphisms in a pretreatment CT-based radiomic model for predicting platinum resistance in ovarian cancer treatment.
Yi X, Liu Y, Zhou B, Xiang W, Deng A, Fu Y, Zhao Y, Ouyang Q, Liu Y, Sun Z, Zhang K, Li X, Zeng F, Zhou H, Chen BT. Yi X, et al. Biomed Pharmacother. 2021 Jan;133:111013. doi: 10.1016/j.biopha.2020.111013. Epub 2020 Nov 20. Biomed Pharmacother. 2021. PMID: 33227705 - An integrated genomic-based approach to individualized treatment of patients with advanced-stage ovarian cancer.
Dressman HK, Berchuck A, Chan G, Zhai J, Bild A, Sayer R, Cragun J, Clarke J, Whitaker RS, Li L, Gray J, Marks J, Ginsburg GS, Potti A, West M, Nevins JR, Lancaster JM. Dressman HK, et al. J Clin Oncol. 2007 Feb 10;25(5):517-25. doi: 10.1200/JCO.2006.06.3743. J Clin Oncol. 2007. PMID: 17290060 Retracted. - Checkpoint kinase 2 (Chk2) supports sensitivity to platinum-based treatment in high grade serous ovarian cancer.
Alkema NG, Tomar T, van der Zee AG, Everts M, Meersma GJ, Hollema H, de Jong S, van Vugt MA, Wisman GB. Alkema NG, et al. Gynecol Oncol. 2014 Jun;133(3):591-8. doi: 10.1016/j.ygyno.2014.03.557. Epub 2014 Mar 20. Gynecol Oncol. 2014. PMID: 24657486 - The contribution of copper efflux transporters ATP7A and ATP7B to chemoresistance and personalized medicine in ovarian cancer.
Lukanović D, Herzog M, Kobal B, Černe K. Lukanović D, et al. Biomed Pharmacother. 2020 Sep;129:110401. doi: 10.1016/j.biopha.2020.110401. Epub 2020 Jun 20. Biomed Pharmacother. 2020. PMID: 32570116 Review. - A systematic literature review assessing if genetic biomarkers are predictors for platinum-based chemotherapy response in ovarian cancer patients.
Phillips-Chavez C, Watson M, Coward J, Schloss J. Phillips-Chavez C, et al. Eur J Clin Pharmacol. 2020 Aug;76(8):1059-1074. doi: 10.1007/s00228-020-02874-4. Epub 2020 May 22. Eur J Clin Pharmacol. 2020. PMID: 32440721
Cited by
- Mass spectrometry-based proteomics techniques and their application in ovarian cancer research.
Swiatly A, Plewa S, Matysiak J, Kokot ZJ. Swiatly A, et al. J Ovarian Res. 2018 Oct 1;11(1):88. doi: 10.1186/s13048-018-0460-6. J Ovarian Res. 2018. PMID: 30270814 Free PMC article. Review. - Histopathology and proteomics are synergistic for High-Grade Serous Ovarian Cancer platinum response prediction.
Kilim O, Olar A, Biricz A, Madaras L, Pollner P, Szállási Z, Sztupinszki Z, Csabai I. Kilim O, et al. medRxiv [Preprint]. 2024 Jun 3:2024.06.01.24308293. doi: 10.1101/2024.06.01.24308293. medRxiv. 2024. PMID: 38883738 Free PMC article. Updated. Preprint. - Omics AnalySIs System for PRecision Oncology (OASISPRO): a web-based omics analysis tool for clinical phenotype prediction.
Yu KH, Fitzpatrick MR, Pappas L, Chan W, Kung J, Snyder M. Yu KH, et al. Bioinformatics. 2018 Jan 15;34(2):319-320. doi: 10.1093/bioinformatics/btx572. Bioinformatics. 2018. PMID: 28968749 Free PMC article. - A deep neural network approach to predicting clinical outcomes of neuroblastoma patients.
Tranchevent LC, Azuaje F, Rajapakse JC. Tranchevent LC, et al. BMC Med Genomics. 2019 Dec 20;12(Suppl 8):178. doi: 10.1186/s12920-019-0628-y. BMC Med Genomics. 2019. PMID: 31856829 Free PMC article. - The Case for Proteomics and Phospho-Proteomics in Personalized Cancer Medicine.
Doll S, Gnad F, Mann M. Doll S, et al. Proteomics Clin Appl. 2019 Mar;13(2):e1800113. doi: 10.1002/prca.201800113. Proteomics Clin Appl. 2019. PMID: 30790462 Free PMC article. Review.
References
- Siegel R; Ma J; Zou Z; Jemal A Cancer statistics, 2014. Ca-Cancer J. Clin. 2014, 64,9–29. - PubMed
- What are the key statistics about ovarian cancer? 2014. http://www.cancer.org/cancer/ovariancancer/detailedguide/ovarian-cancer-... (accessed December 11, 2014).
- Goff BA; Mandel LS; Melancon CH; Muntz HG Frequency of symptoms of ovarian cancer in women presenting to primary care clinics. Jama 2004, 291, 2705–12. - PubMed
- Olson SH; Mignone L; Nakraseive C; Caputo TA; Barakat RR; Harlap S Symptoms of ovarian cancer. Obstet. Gynecol. 2001, 98, 212–7. - PubMed
- Goff BA; Mandel L; Muntz HG; Melancon CH Ovarian carcinoma diagnosis. Cancer 2000, 89, 2068–75. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous