Autoubiquitination of the Hrd1 Ligase Triggers Protein Retrotranslocation in ERAD - PubMed (original) (raw)
Autoubiquitination of the Hrd1 Ligase Triggers Protein Retrotranslocation in ERAD
Ryan D Baldridge et al. Cell. 2016.
Abstract
Misfolded proteins of the ER are retrotranslocated to the cytosol, where they are polyubiquitinated, extracted from the membrane, and degraded by the proteasome. To investigate how the ER-associated Degradation (ERAD) machinery can accomplish retrotranslocation of a misfolded luminal protein domain across a lipid bilayer, we have reconstituted retrotranslocation with purified S. cerevisiae proteins, using proteoliposomes containing the multi-spanning ubiquitin ligase Hrd1. Retrotranslocation of the luminal domain of a membrane-spanning substrate is triggered by autoubiquitination of Hrd1. Substrate ubiquitination is a subsequent event, and the Cdc48 ATPase that completes substrate extraction from the membrane is not required for retrotranslocation. Ubiquitination of lysines in Hrd1's RING-finger domain is required for substrate retrotranslocation in vitro and for ERAD in vivo. Our results suggest that Hrd1 forms a ubiquitin-gated protein-conducting channel.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
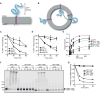
Figure 1. Membrane-bound ERAD substrates of the Hrd1 ubiquitin ligase
(A) Scheme of CPY*-TM, a fusion of the misfolded ER luminal substrate CPY* (blue) with a transmembrane (TM) segment derived from the multi-spanning membrane protein Pdr5 (purple). (B) CPY*-TM is reconstituted into proteoliposomes in both orientations. Retrotranslocation is defined as the movement of a segment of the luminal domain from the inside to the outside of the vesicles (arrow). (C) The degradation of CPY*-TM with a C-terminal HA tag was tested after addition of cycloheximide to wild-type S. cerevisiae cells or strains lacking the indicated ERAD components. At each time point, proteins were analyzed by SDS-PAGE and immunoblotting with HA antibodies. Full-length protein and a degradation product were observed (Figure S1A). The quantification shows the sum of the intensity of both bands. Data are represented as mean +/− SEM from at least three experiments. (D) As in (C), but with CPY*-TM2, which contains the TM segment from the single-spanning membrane protein Spt23 (see also Figure S1B). (E) Purified, DyLight800-labeled CPY*-TM, CPY*-TM2, or CPY* (each 100 nM) was incubated with different concentrations of purified Hrd1-SBP immobilized on streptavidin beads. The bound material was analyzed by SDS-PAGE and fluorescence scanning. For comparison, the binding of fusions of wild-type CPY with TM or TM2 (CPY-TM and CPY-TM2, respectively) was tested (dotted lines). The apparent affinities for CPY*, CPY*-TM, CPY*-TM2, CPY-TM, and CPY-TM2 were estimated to be 20 nM, 40 nM, 30 nM, 120 nM, and 110 nM, respectively. (F) The indicated DyLight800-labeled proteins were incubated with Hrd1 and ubiquitination machinery in detergent. Controls were performed in the absence of ATP. The samples were analyzed by SDS-PAGE and fluorescence scanning. (G) Quantification of the experiments in (F). See also Figure S1.
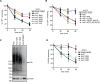
Figure 2. Hrd1 polyubiquitinates CPY*-TM in reconstituted proteoliposomes
(A) Scheme of the reconstitution protocol. CPY*-TM (red) was mixed with phospholipids totally solubilized in Triton X-100 and the detergent was removed. Then, the vesicles were partially solubilized in the detergent DMNG, Hrd1 (blue) was added, and the detergent was removed. (B) Reconstituted proteoliposomes (PLsomes) containing fluorescently labeled CPY*-TM (red; DyLight680) and Hrd1 (blue; DyLight800) were subjected to flotation in a glycerol gradient. Fractions were analyzed by SDS-PAGE and fluorescence scanning (bottom (B) is fraction 1 and top (T) is fraction 6). (C) PLsomes containing amine-labeled Hrd1 (DyLight800) with a unique TEV-cleavage site near its C-terminus were incubated with TEV protease in the presence or absence of Triton X-100 (X100). Controls were performed with 3C protease, a related protease that should not cleave Hrd1. The samples were analyzed by SDS-PAGE and fluorescence scanning. Note that all Hrd1 molecules have their C-terminus on the outside of the vesicles. (D) PLsomes as in (B) were incubated with ubiquitination machinery for different time periods. Controls were performed in the absence of the indicated components. The samples were analyzed by SDS-PAGE and fluorescence scanning at two different wavelengths. (E) PLsomes containing fluorescently labeled CPY*-TM (blue; DyLight800) and unlabeled Hrd1 were mixed with PLsomes containing labeled CPY*-TM (red; DyLight680), but lacking Hrd1. Ubiquitination machinery was added for different time periods, with a control lacking ATP. Note that substrate is modified only if co-reconstituted with Hrd1. (F) PLsomes containing Hrd1 and DyLight800-labeled CPY*-TM were incubated with CPY* and ubiquitination machinery for different time periods. Note that CPY* remains unmodified. See also Figure S2.
Figure 3. Hrd1 retrotranslocates the misfolded CPY* domain of CPY*-TM
(A) Specific labeling of differently oriented substrate molecules. CPY*-TM was labeled by sortase with a fluorescent probe (blue; DyLight800) at the C-terminus, and incorporated into proteoliposomes (PLsomes) in both orientations. The vesicles were then incubated with sortase and peptide coupled to another fluorescent dye (red; DyLight680), switching the color of substrate molecules that have their C-terminus accessible. Finally, Hrd1 was incorporated into the vesicles. (B) The orientation of substrate molecules in reconstituted PLsomes was tested by addition of different concentrations of the cysteine-modifying reagent Mal-PEG. Controls were performed in the presence of detergent. The samples were analyzed by SDS-PAGE and fluorescence scanning. Note that the CPY* domain of the red molecules can be modified only after addition of detergent. (C) The orientation of substrate molecules was tested in PLsomes containing inactive Hrd1 ligase (Hrd1 C399S). Substrate lysine ubiquitination by the ligase Rsp5 was followed over time. Controls were performed in the presence of detergent. Note that the CPY* domain of the red molecules is modified by Rsp5 only after addition of detergent. (D) PLsomes containing wild-type Hrd1 and CPY*-TM in both orientations were incubated with ubiquitination machinery for different time periods in the presence or absence of detergent. Note that the CPY* domain of the red molecules is modified even in the absence of detergent. (E) As in (D), but with PLsomes containing either wild-type Hrd1 or a mutant that lacks TMs 3-6 (mini-Hrd1). Note that mini-Hrd1 is active in polyubiqitination, but does not support modification of the CPY* domain of the red molecules unless detergent is added. (F) Scheme of the fluorescence quenching retrotranslocation assay. PLsomes were generated as in (D), but CPY*-TM also contained an additional lysine-attached fluorescent dye on the CPY* domain (Alexa488; indicated by green domain) to which a quenching antibody (shown in gray) is available. The antibody quenched the fluorescence of substrate molecules that had their CPY* domain on the outside of the vesicles (gray domains). Substrate molecules of the opposite orientation can only be quenched after retrotranslocation of their CPY* domain. (G) PLsomes as in (F) were incubated with antibody and ubiquitination machinery in the absence of ATP. The sample was placed in a fluorescence plate reader, ATP was added (open arrow), and the decrease in fluorescence was followed over time. A control was performed by addition of Triton X-100 (X100) before the reaction (open arrow). Each sample received Triton X-100 after the reaction (closed arrow). (H) PLsomes were generated with catalytically inactive Hrd1 (C399S) and CPY*-TM that was labeled at the C-terminus with Dylight 800 (blue) and at lysines in the CPY* domain with Alexa488 (green). Retrotranslocation was assayed as in (G). See also Figure S3.
Figure 4. Hrd1 retrotranslocates a lysine-free substrate in vitro and in vivo
(A) The degradation of lysine-free CPY* was tested after addition of cycloheximide in wild-type S. cerevisiae cells and strains lacking the indicated ERAD components. At each time point, proteins were analyzed by SDS-PAGE and immunoblotting with HA antibodies (see also Figure S4A). Data are represented as mean +/− SEM from at least three experiments. (B) As in (A), but with lysine-free CPY*-TM. Full-length protein and a C-terminal degradation product were observed (Figure S4B). The quantification shows the sum of both. (C) Lysine-free CPY*-TM (noK) was incubated with Hrd1 and ubiquitination machinery for different time periods. Where indicated, the substrate was pre-modified with the cysteine-modifying reagent N-ethylmaleimide (NEM). The samples were analyzed by non-reducing (Non-Red) or reducing (Red) SDS-PAGE. Shown is the quantification of non-ubiquitinated Hrd1 and substrate (see also Figure S4C). (D) The retrotranslocation of CPY*-TM (no K) was tested with the fluorescence quenching assay as in Figure 3F. Note that the lysine-free CPY* domain gradually becomes accessible to the quenching antibodies. (E) The samples in (D) were analyzed for substrate polyubiquitination by SDS-PAGE and fluorescence scanning. (F) The fraction of retrotranslocated CPY* domain (black line), as assessed by fluorescence quenching in (D), is compared with the fraction that remains non-ubiquitinated (red line) in the same reaction (E). The blue line shows the fraction of unmodified substrate molecules that have their CPY* domain on the outside of the vesicles. See also Figure S4.
Figure 5. Autoubiquitination of Hrd1 is required for retrotranslocation
(A) DyLight800-labeled CPY*-TM was incubated with different concentrations of unmodified Hrd1, pre-polyubiquitinated Hrd1 (poly-Ub), or polyubiquitinated Hrd1 treated with the de-ubiquitinating enzyme Usp2 (de-Ub). After incubation with streptavidin beads, the amount of bound material was analyzed by SDS-PAGE and fluorescence scanning. Note that modified Hrd1 has a higher affinity for substrate. (B) As in (A), but Hrd1 was also pre-modified with mono-, di- or tetra-ubiquitin. (C) As in (A), but Hrd1 variants were used, which contained lysine-to-arginine substitutions in either the transmembrane domain (RKK), the RING-finger domain (KRK), or the C-terminal tail (KKR). In each case, Hrd1 was tested with and without polyubiquitination. Note that the ubiquitination-mediated increase of substrate affinity is blunted for the KRK mutant. (D) Proteoliposomes containing substrate in both orientations and one of the Hrd1 mutants described in (C) were incubated with ubiquitination machinery for different time periods in the presence or absence of detergent. The samples were analyzed by SDS-PAGE and fluorescence scanning. Note that the CPY* domain of the red molecules is not efficiently retrotranslocated by the KRK mutant. (E) Quantification of the fluorescence at 800 nm in (D). (F) Quantification of the fluorescence at 680 nm in (D). (G) Retrotranslocation of CPY*-TM was tested with the fluorescence quenching assay as in Figure 3H, but with Hrd1 carrying lysine to arginine substitutions in the RING domain (KRK mutant). (H) As in (G), but with Hrd1 containing lysines only in the RING domain (Hrd1 RKR). See also Figure S5.
Figure 6. Hrd1 autoubiquitination affects ERAD
(A) The degradation of CPY* was followed in S. cerevisiae cells lacking Hrd1 (hrd1Δ), which contained either an empty vector or expressed Hrd1 mutants with lysine-to-arginine mutations from a single-copy plasmid. At each time point after cycloheximide addition, proteins were analyzed by SDS-PAGE and immunoblotting with HA antibodies (see also Figure S6A). Data are represented as mean +/− SEM from at least three experiments. Note that CPY* is not degraded in the KRK mutant. (B) As in (A), but with Hrd1 mutants carrying the indicated lysine to arginine substitutions in the RING-finger domain. Note that a Hrd1 mutant carrying mutations in three lysines (373, 387, 407) is inactive in ERAD of CPY*. (C) Wild-type Hrd1 or the indicated Hrd1 mutants with a His10-tag at their C-termini were expressed in hrd1Δ cells from a single-copy plasmid. The proteins were purified with IMAC beads under denaturing conditions, separated by SDS-PAGE, and subjected to immunoblotting with Hrd1 and ubiquitin antibodies. Note that the RKR mutant is polyubiquitinated. (D) As in (A), but with the ERAD-L substrate KHN. (See also Figure S6G) See also Figure S6.
Figure 7. A model for Hrd1-mediated retrotranslocation
The scheme shows different stages of basic ERAD. Stage 1 shows the ubiquitin ligase Hrd1 and a single-spanning ERAD substrate containing a misfolded luminal domain. In stage 2, a segment of the misfolded domain interacts with the Hrd1 ligase. In stage 3, Hrd1 is autoubiquitinated, the substrate is inserted as a loop, and can slide back and forth inside the Hrd1 channel. In stage 4, the substrate is polyubiquitinated, which prevents back-sliding of the polypeptide into the ER lumen. In stage 5, the Cdc48 ATPase complex extracts the polyubiquitinated substrate from the ER membrane. See also Table S1.
Comment in
- Ever HRD a ubiquitin-gated channel?
Zhang T, Ye Y. Zhang T, et al. Cell Res. 2016 Oct;26(10):1075-1076. doi: 10.1038/cr.2016.92. Epub 2016 Aug 5. Cell Res. 2016. PMID: 27491350 Free PMC article.
Similar articles
- Key steps in ERAD of luminal ER proteins reconstituted with purified components.
Stein A, Ruggiano A, Carvalho P, Rapoport TA. Stein A, et al. Cell. 2014 Sep 11;158(6):1375-1388. doi: 10.1016/j.cell.2014.07.050. Cell. 2014. PMID: 25215493 Free PMC article. - Cycles of autoubiquitination and deubiquitination regulate the ERAD ubiquitin ligase Hrd1.
Peterson BG, Glaser ML, Rapoport TA, Baldridge RD. Peterson BG, et al. Elife. 2019 Nov 12;8:e50903. doi: 10.7554/eLife.50903. Elife. 2019. PMID: 31713515 Free PMC article. - A Cdc48 "Retrochaperone" Function Is Required for the Solubility of Retrotranslocated, Integral Membrane Endoplasmic Reticulum-associated Degradation (ERAD-M) Substrates.
Neal S, Mak R, Bennett EJ, Hampton R. Neal S, et al. J Biol Chem. 2017 Feb 24;292(8):3112-3128. doi: 10.1074/jbc.M116.770610. Epub 2017 Jan 11. J Biol Chem. 2017. PMID: 28077573 Free PMC article. - Mechanistic insights into ER-associated protein degradation.
Wu X, Rapoport TA. Wu X, et al. Curr Opin Cell Biol. 2018 Aug;53:22-28. doi: 10.1016/j.ceb.2018.04.004. Epub 2018 Apr 30. Curr Opin Cell Biol. 2018. PMID: 29719269 Free PMC article. Review. - Protein Quality Control of the Endoplasmic Reticulum and Ubiquitin-Proteasome-Triggered Degradation of Aberrant Proteins: Yeast Pioneers the Path.
Berner N, Reutter KR, Wolf DH. Berner N, et al. Annu Rev Biochem. 2018 Jun 20;87:751-782. doi: 10.1146/annurev-biochem-062917-012749. Epub 2018 Feb 2. Annu Rev Biochem. 2018. PMID: 29394096 Review.
Cited by
- Yeast derlin Dfm1 employs a chaperone-like function to resolve misfolded membrane protein stress.
Kandel R, Jung J, Syau D, Kuo T, Songster L, Horn C, Chapman C, Aguayo A, Duttke S, Benner C, Neal SE. Kandel R, et al. PLoS Biol. 2023 Jan 23;21(1):e3001950. doi: 10.1371/journal.pbio.3001950. eCollection 2023 Jan. PLoS Biol. 2023. PMID: 36689475 Free PMC article. - Ubiquitin-dependent protein degradation at the endoplasmic reticulum and nuclear envelope.
Mehrtash AB, Hochstrasser M. Mehrtash AB, et al. Semin Cell Dev Biol. 2019 Sep;93:111-124. doi: 10.1016/j.semcdb.2018.09.013. Epub 2018 Oct 9. Semin Cell Dev Biol. 2019. PMID: 30278225 Free PMC article. Review. - Accumulated precursors of specific GPI-anchored proteins upregulate GPI biosynthesis with ARV1.
Liu YS, Wang Y, Zhou X, Zhang L, Yang G, Gao XD, Murakami Y, Fujita M, Kinoshita T. Liu YS, et al. J Cell Biol. 2023 May 1;222(5):e202208159. doi: 10.1083/jcb.202208159. Epub 2023 Feb 24. J Cell Biol. 2023. PMID: 36828365 Free PMC article. - Disposing of misfolded ER proteins: A troubled substrate's way out of the ER.
Oikonomou C, Hendershot LM. Oikonomou C, et al. Mol Cell Endocrinol. 2020 Jan 15;500:110630. doi: 10.1016/j.mce.2019.110630. Epub 2019 Oct 24. Mol Cell Endocrinol. 2020. PMID: 31669350 Free PMC article. Review. - The degradation-promoting roles of deubiquitinases Ubp6 and Ubp3 in cytosolic and ER protein quality control.
Wu H, Ng DTW, Cheong I, Matsudaira P. Wu H, et al. PLoS One. 2020 May 13;15(5):e0232755. doi: 10.1371/journal.pone.0232755. eCollection 2020. PLoS One. 2020. PMID: 32401766 Free PMC article.
References
- Ben-Saadon R, Zaaroor D, Ziv T, Ciechanover A. The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol Cell. 2006;24:701–711. - PubMed
- Bordallo J, Wolf DH. A RING-H2 finger motif is essential for the function of Der3/Hrd1 in endoplasmic reticulum associated protein degradation in the yeast Saccharomyces cerevisiae. FEBS Lett. 1999;448:244–248. - PubMed
- Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309:127–130. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases