Mammal madness: is the mammal tree of life not yet resolved? - PubMed (original) (raw)
Review
Mammal madness: is the mammal tree of life not yet resolved?
Nicole M Foley et al. Philos Trans R Soc Lond B Biol Sci. 2016.
Abstract
Most molecular phylogenetic studies place all placental mammals into four superordinal groups, Laurasiatheria (e.g. dogs, bats, whales), Euarchontoglires (e.g. humans, rodents, colugos), Xenarthra (e.g. armadillos, anteaters) and Afrotheria (e.g. elephants, sea cows, tenrecs), and estimate that these clades last shared a common ancestor 90-110 million years ago. This phylogeny has provided a framework for numerous functional and comparative studies. Despite the high level of congruence among most molecular studies, questions still remain regarding the position and divergence time of the root of placental mammals, and certain 'hard nodes' such as the Laurasiatheria polytomy and Paenungulata that seem impossible to resolve. Here, we explore recent consensus and conflict among mammalian phylogenetic studies and explore the reasons for the remaining conflicts. The question of whether the mammal tree of life is or can be ever resolved is also addressed.This article is part of the themed issue 'Dating species divergences using rocks and clocks'.
Keywords: coalesence; fossils; molecules; phylogenomics; supermatrix; time-tree.
© 2016 The Author(s).
Figures
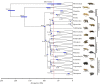
Figure 1.
Timetree based on mcmctree supports a long fuse model of mammalian diversification [4,5]. Divergence time estimates were obtained under a GTR + Γ model of sequence evolution, 99 calibrated nodes (see electronic supplementary material table S2 for details), and autocorrelated rates of evolution. Divergence time estimates for clades of interest are shown at each node, with the 95% CI denoted by blue bars. The dataset included 26 tree of life gene fragments [4] and was expanded to include 286 taxa (see electronic supplementary material, table S1 for taxa, gene fragments and accession numbers). GenBank sequences for single genes were supplemented with available genome data that were mined using BLAST searches. The tree was estimated using RAxML [99] on the CIPRES web server [100] under a fully partitioned model where each partition had its own GTR + Γ model of sequence evolution. Paintings by Carl Buell. (Online version in colour.)
Similar articles
- Afrotheria.
Springer MS. Springer MS. Curr Biol. 2022 Mar 14;32(5):R205-R210. doi: 10.1016/j.cub.2022.02.001. Curr Biol. 2022. PMID: 35290765 - Molecular phylogeny of living xenarthrans and the impact of character and taxon sampling on the placental tree rooting.
Delsuc F, Scally M, Madsen O, Stanhope MJ, de Jong WW, Catzeflis FM, Springer MS, Douzery EJ. Delsuc F, et al. Mol Biol Evol. 2002 Oct;19(10):1656-71. doi: 10.1093/oxfordjournals.molbev.a003989. Mol Biol Evol. 2002. PMID: 12270893 - Molecular phylogenetics and the origins of placental mammals.
Murphy WJ, Eizirik E, Johnson WE, Zhang YP, Ryder OA, O'Brien SJ. Murphy WJ, et al. Nature. 2001 Feb 1;409(6820):614-8. doi: 10.1038/35054550. Nature. 2001. PMID: 11214319 - The new framework for understanding placental mammal evolution.
Asher RJ, Bennett N, Lehmann T. Asher RJ, et al. Bioessays. 2009 Aug;31(8):853-64. doi: 10.1002/bies.200900053. Bioessays. 2009. PMID: 19582725 Review. - The chromosomes of Afrotheria and their bearing on mammalian genome evolution.
Svartman M, Stanyon R. Svartman M, et al. Cytogenet Genome Res. 2012;137(2-4):144-53. doi: 10.1159/000341387. Epub 2012 Aug 3. Cytogenet Genome Res. 2012. PMID: 22868637 Review.
Cited by
- Chromosome-level genome and population genomics of the intermediate horseshoe bat ( Rhinolophus affinis) reveal the molecular basis of virus tolerance in Rhinolophus and echolocation call frequency variation.
Zhao L, Yuan J, Wang G, Jing H, Huang C, Xu L, Xu X, Sun T, Chen W, Mao X, Li G. Zhao L, et al. Zool Res. 2024 Sep 18;45(5):1147-1160. doi: 10.24272/j.issn.2095-8137.2024.027. Zool Res. 2024. PMID: 39257377 Free PMC article. - SMA20/PMIS2 Is a Rapidly Evolving Sperm Membrane Alloantigen with Possible Species-Divergent Function in Fertilization.
Cormier N, Worsham AE, Rich KA, Hardy DM. Cormier N, et al. Int J Mol Sci. 2024 Mar 25;25(7):3652. doi: 10.3390/ijms25073652. Int J Mol Sci. 2024. PMID: 38612464 Free PMC article. - Translating the Timing of Developmental Benchmarks in Short-Tailed Opossums (Monodelphisdomestica) to Facilitate Comparisons with Commonly Used Rodent Models.
Bresee C, Litman-Cleper J, Clayton CJ, Krubitzer L. Bresee C, et al. Brain Behav Evol. 2024;99(2):69-85. doi: 10.1159/000538524. Epub 2024 Mar 25. Brain Behav Evol. 2024. PMID: 38527443 - The evolution of menopause in toothed whales.
Ellis S, Franks DW, Nielsen MLK, Weiss MN, Croft DP. Ellis S, et al. Nature. 2024 Mar;627(8004):579-585. doi: 10.1038/s41586-024-07159-9. Epub 2024 Mar 13. Nature. 2024. PMID: 38480878 Free PMC article. - Geologically calibrated mammalian tree and its correlation with global events, including the emergence of humans.
Osozawa S. Osozawa S. Ecol Evol. 2023 Dec 19;13(12):e10827. doi: 10.1002/ece3.10827. eCollection 2023 Dec. Ecol Evol. 2023. PMID: 38116126 Free PMC article.
References
- Wilson DE, Reeder DM. 2005. Mammal species of the world: a taxonomic and geographic reference. Baltimore, MD: Johns Hopkins University Press.
- McKenna MC, Bell SK. 1997. Classification of mammals. New York, NY: Columbia University Press.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources