16S rRNA gene sequencing of mock microbial populations- impact of DNA extraction method, primer choice and sequencing platform - PubMed (original) (raw)
16S rRNA gene sequencing of mock microbial populations- impact of DNA extraction method, primer choice and sequencing platform
Fiona Fouhy et al. BMC Microbiol. 2016.
Abstract
Background: Next-generation sequencing platforms have revolutionised our ability to investigate the microbiota composition of complex environments, frequently through 16S rRNA gene sequencing of the bacterial component of the community. Numerous factors, including DNA extraction method, primer sequences and sequencing platform employed, can affect the accuracy of the results achieved. The aim of this study was to determine the impact of these three factors on 16S rRNA gene sequencing results, using mock communities and mock community DNA.
Results: The use of different primer sequences (V4-V5, V1-V2 and V1-V2 degenerate primers) resulted in differences in the genera and species detected. The V4-V5 primers gave the most comparable results across platforms. The three Ion PGM primer sets detected more of the 20 mock community species than the equivalent MiSeq primer sets. Data generated from DNA extracted using the 2 extraction methods were very similar.
Conclusions: Microbiota compositional data differed depending on the primers and sequencing platform that were used. The results demonstrate the risks in comparing data generated using different sequencing approaches and highlight the merits of choosing a standardised approach for sequencing in situations where a comparison across multiple sequencing runs is required.
Keywords: 16S rRNA; Bias; DNA extraction; Gut microbiota; Ion PGM; MiSeq; Mock communities; Next-generation sequencing.
Figures
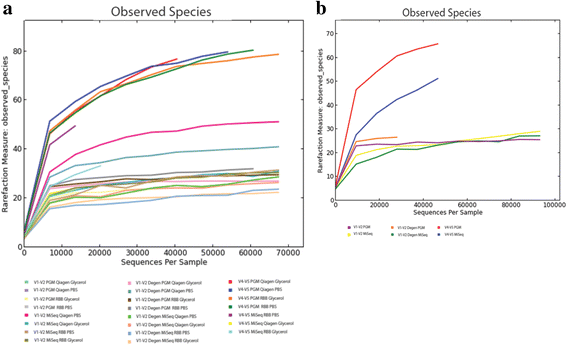
Fig. 1
Rarefaction curves based on sample ID and number of observed species for mock cells (a) and mock DNA samples (b). Curves are approaching or are horizontal with the x axis indicating that additional sequencing would not yield additional novel data
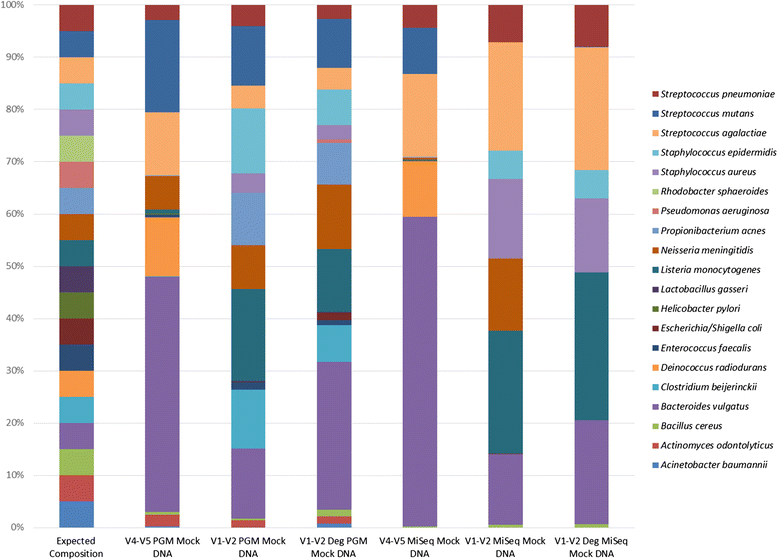
Fig. 2
Percentage relative abundance of expected species (n = 20) detected in mock community DNA based on sequencing platform and primer set used
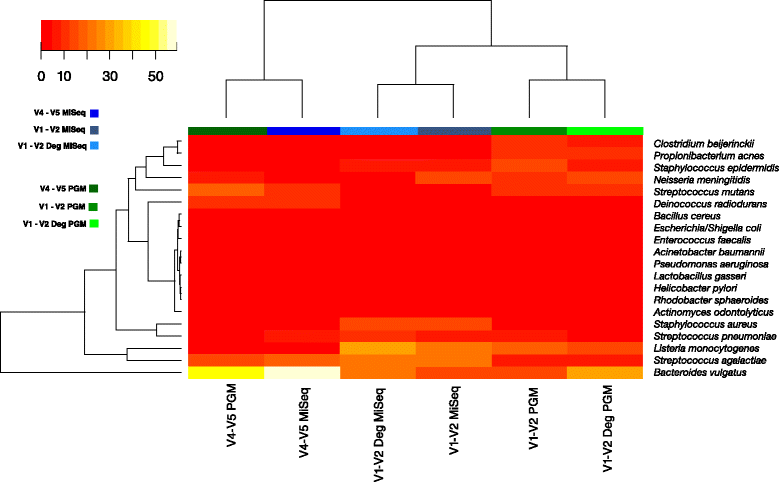
Fig. 3
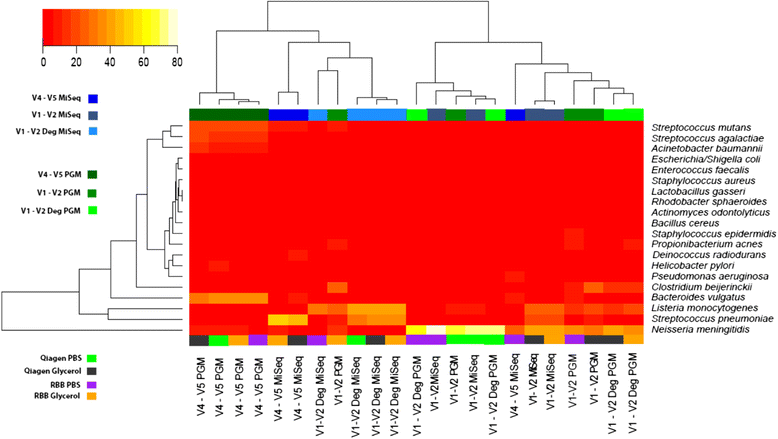
Heat map of species abundance. Only the 20 expected taxa from mock DNA (HM-782D) were included. Hierarchical clustering was performed using hclusing default parameters (complete linkage). The blue colours represent samples sequenced on MiSeq platform while green represent the Ion PGM
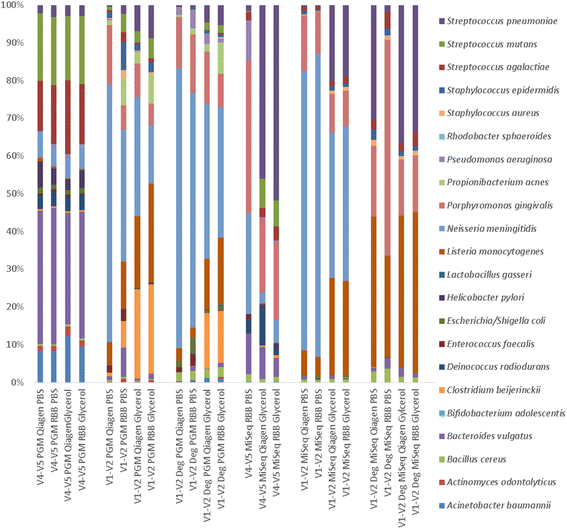
Fig. 4
Percentage relative abundance of expected species based on extraction procedure
Fig. 5
Heat map of species abundance by sequencer and extraction method for mock community cells. Only expected taxa were included and hierarchical clustering was performed using hclust default parameters (complete linkage). The top colour legend depicts the sequencing technology and primer set. The blue colours represent samples sequenced on MiSeq platform while green represent the Ion PGM. The bottom legend represents the samples and extraction method
Similar articles
- Optimisation of methods for bacterial skin microbiome investigation: primer selection and comparison of the 454 versus MiSeq platform.
Castelino M, Eyre S, Moat J, Fox G, Martin P, Ho P, Upton M, Barton A. Castelino M, et al. BMC Microbiol. 2017 Jan 21;17(1):23. doi: 10.1186/s12866-017-0927-4. BMC Microbiol. 2017. PMID: 28109256 Free PMC article. - Primer, Pipelines, Parameters: Issues in 16S rRNA Gene Sequencing.
Abellan-Schneyder I, Matchado MS, Reitmeier S, Sommer A, Sewald Z, Baumbach J, List M, Neuhaus K. Abellan-Schneyder I, et al. mSphere. 2021 Feb 24;6(1):e01202-20. doi: 10.1128/mSphere.01202-20. mSphere. 2021. PMID: 33627512 Free PMC article. - A 16S rRNA gene sequencing and analysis protocol for the Illumina MiniSeq platform.
Pichler M, Coskun ÖK, Ortega-Arbulú AS, Conci N, Wörheide G, Vargas S, Orsi WD. Pichler M, et al. Microbiologyopen. 2018 Dec;7(6):e00611. doi: 10.1002/mbo3.611. Epub 2018 Mar 25. Microbiologyopen. 2018. PMID: 29575567 Free PMC article. - Generating amplicon reads for microbial community assessment with next-generation sequencing.
Gołębiewski M, Tretyn A. Gołębiewski M, et al. J Appl Microbiol. 2020 Feb;128(2):330-354. doi: 10.1111/jam.14380. Epub 2019 Aug 1. J Appl Microbiol. 2020. PMID: 31299126 Review. - Profiling of Oral Bacterial Communities.
Wade WG, Prosdocimi EM. Wade WG, et al. J Dent Res. 2020 Jun;99(6):621-629. doi: 10.1177/0022034520914594. Epub 2020 Apr 14. J Dent Res. 2020. PMID: 32286907 Free PMC article. Review.
Cited by
- Assessment of Skimmed Milk Flocculation for Bacterial Enrichment from Water Samples, and Benchmarking of DNA Extraction and 16S rRNA Databases for Metagenomics.
Donchev D, Stoikov I, Diukendjieva A, Ivanov IN. Donchev D, et al. Int J Mol Sci. 2024 Oct 8;25(19):10817. doi: 10.3390/ijms251910817. Int J Mol Sci. 2024. PMID: 39409144 Free PMC article. - Stability of temperate coral Astrangia poculata microbiome is reflected across different sequencing methodologies.
Goldsmith DB, Pratte ZA, Kellogg CA, Snader SE, Sharp KH. Goldsmith DB, et al. AIMS Microbiol. 2019 Mar 1;5(1):62-76. doi: 10.3934/microbiol.2019.1.62. eCollection 2019. AIMS Microbiol. 2019. PMID: 31384703 Free PMC article. - Anthropogenic Intensity-Determined Assembly and Network Stability of Bacterioplankton Communities in the Le'an River.
Wu B, Wang P, Devlin AT, She Y, Zhao J, Xia Y, Huang Y, Chen L, Zhang H, Nie M, Ding M. Wu B, et al. Front Microbiol. 2022 May 4;13:806036. doi: 10.3389/fmicb.2022.806036. eCollection 2022. Front Microbiol. 2022. PMID: 35602050 Free PMC article. - Metagenomics reveals novel microbial signatures of farm exposures in house dust.
Wang Z, Dalton KR, Lee M, Parks CG, Beane Freeman LE, Zhu Q, González A, Knight R, Zhao S, Motsinger-Reif AA, London SJ. Wang Z, et al. medRxiv [Preprint]. 2023 Apr 12:2023.04.07.23288301. doi: 10.1101/2023.04.07.23288301. medRxiv. 2023. PMID: 37090637 Free PMC article. Updated. Preprint. - Helminth Microbiota Profiling Using Bacterial 16S rRNA Gene Amplicon Sequencing: From Sampling to Sequence Data Mining.
Formenti F, Rinaldi G, Cantacessi C, Cortés A. Formenti F, et al. Methods Mol Biol. 2021;2369:263-298. doi: 10.1007/978-1-0716-1681-9_15. Methods Mol Biol. 2021. PMID: 34313994
References
- Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, Murphy B, Ross RP, Fitzgerald GF, Stanton C. High-throughput sequencing reveals the incomplete, short-term, recovery of the infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamycin. Antimicrob Agents Chemother. 2012;56(11):5811–20. - PMC - PubMed
- Murphy E, Cotter P, Healy S, Marques T, O’Sullivan O, Fouhy F, Clarke S, O'Toole P, Quigley E, Stanton C. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59(12):1635–42. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources