Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells - PubMed (original) (raw)
Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells
Benjamin P Kleinstiver et al. Nat Biotechnol. 2016 Aug.
Abstract
The activities and genome-wide specificities of CRISPR-Cas Cpf1 nucleases are not well defined. We show that two Cpf1 nucleases from Acidaminococcus sp. BV3L6 and Lachnospiraceae bacterium ND2006 (AsCpf1 and LbCpf1, respectively) have on-target efficiencies in human cells comparable with those of the widely used Streptococcus pyogenes Cas9 (SpCas9). We also report that four to six bases at the 3' end of the short CRISPR RNA (crRNA) used to program Cpf1 nucleases are insensitive to single base mismatches, but that many of the other bases in this region of the crRNA are highly sensitive to single or double substitutions. Using GUIDE-seq and targeted deep sequencing analyses performed with both Cpf1 nucleases, we were unable to detect off-target cleavage for more than half of 20 different crRNAs. Our results suggest that AsCpf1 and LbCpf1 are highly specific in human cells.
Figures
Figure 1
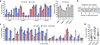
On-target indel mutation percentages induced by Cpf1 nucleases in human cells. (a) Endogenous gene percent modification induced by AsCpf1 and LbCpf1 at 19 endogenous human gene sites (left panel, determined by T7E1 assay). Target sites were chosen in amplicons known to be efficiently modified by SpCas9 (right panel, and see Supplementary Fig. 1). Error bars, s.e.m.; n = 3. (b) and (c) Matched target sites for AsCpf1, LbCpf1, and SpCas9 that share a common protospacer sequence (panel b) were examined for mutagenesis by AsCpf1, LbCpf1, and SpCas9 nucleases (determined by T7E1 assay; panel c). Error bars, s.e.m.; n = at least 3; ND, not detected. (d) Summary of matched site on-target activities from panel c, with means and 95% confidence intervals shown.
Figure 2
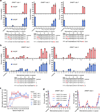
Tolerance of AsCpf1 and LbCpf1 to mismatched or truncated crRNAs. (a, b) Endogenous gene modification by AsCpf1 and LbCpf1 using crRNAs that contain pairs of mismatched bases (panel a) or singly mismatched bases (panel b). Activity determined by T7E1 assay; error bars, s.e.m.; n = 3. (c) Summary of mutagenesis percentages by AsCpf1 and LbCpf1 at 3 different endogenous sites with crRNAs bearing variable length 3’ end truncations or extensions (see also Supplementary Fig. 3a), where activities are normalized to mutagenesis observed when using the canonical 23 nucleotide spacer. Error bars, s.e.m.; n = 3; nt, nucleotide. (d) Activity of AsCpf1 and LbCpf1 when programed with singly mismatched crRNAs either of 23 or 20 nucleotides in length. For clarity, data from panel b is re-presented here. Activity determined by T7E1 assay; error bars, s.e.m.; n = 3.
Figure 3
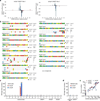
Genome-wide specificities of AsCpf1 and LbCpf1. (a) Relative GUIDE-seq read start mapping position. The first 5’ base of the protospacer adjacent to the PAM is position 1. Reads that start within the protospacer region are colored in blue, reads that originate outside are colored in red. Reads mapping to the plus strand are depicted above the x-axis, reads mapping in the reverse orientation are depicted below. (b) Off-target sites for AsCpf1 and LbCpf1 with 20 different crRNAs, determined using GUIDE-seq in U2OS cells. Mismatched positions in the target sites of off-targets are highlighted in color, and GUIDE-seq read counts shown to the right of the on- and off-target sequences represent a measure of cleavage efficiency at a given site. (c) and (d) Summary of the total number of off-target sites detected by GUIDE-seq in U2OS cells with 20 crRNAs (panel c) and in HEK293 cells with 3 crRNAs (panel d). The off-target sequences and GUIDE-seq read counts for matched site #6 are shown in Supplementary Fig. 5. (e) Scatterplots of mean mutagenesis versus GUIDE-seq read counts analyzed from independent samples. Mutagenesis percentages for AsCpf1 that are not significantly different from negative controls are indicated with triangles. Error bars, s.e.m.; n = 3.
Similar articles
- Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells.
Kim D, Kim J, Hur JK, Been KW, Yoon SH, Kim JS. Kim D, et al. Nat Biotechnol. 2016 Aug;34(8):863-8. doi: 10.1038/nbt.3609. Epub 2016 Jun 6. Nat Biotechnol. 2016. PMID: 27272384 - Cpf1 nucleases demonstrate robust activity to induce DNA modification by exploiting homology directed repair pathways in mammalian cells.
Tóth E, Weinhardt N, Bencsura P, Huszár K, Kulcsár PI, Tálas A, Fodor E, Welker E. Tóth E, et al. Biol Direct. 2016 Sep 14;11:46. doi: 10.1186/s13062-016-0147-0. Biol Direct. 2016. PMID: 27630115 Free PMC article. - Efficient genome editing in wheat using Cas9 and Cpf1 (AsCpf1 and LbCpf1) nucleases.
Kim D, Hager M, Brant E, Budak H. Kim D, et al. Funct Integr Genomics. 2021 Jul;21(3-4):355-366. doi: 10.1007/s10142-021-00782-z. Epub 2021 Mar 12. Funct Integr Genomics. 2021. PMID: 33710467 - Cas9, Cpf1 and C2c1/2/3-What's next?
Nakade S, Yamamoto T, Sakuma T. Nakade S, et al. Bioengineered. 2017 May 4;8(3):265-273. doi: 10.1080/21655979.2017.1282018. Epub 2017 Jan 31. Bioengineered. 2017. PMID: 28140746 Free PMC article. Review. - The Conspicuity of CRISPR-Cpf1 System as a Significant Breakthrough in Genome Editing.
Bayat H, Modarressi MH, Rahimpour A. Bayat H, et al. Curr Microbiol. 2018 Jan;75(1):107-115. doi: 10.1007/s00284-017-1406-8. Epub 2017 Nov 30. Curr Microbiol. 2018. PMID: 29189942 Review.
Cited by
- Leveraging CRISPR gene editing technology to optimize the efficacy, safety and accessibility of CAR T-cell therapy.
Lei T, Wang Y, Zhang Y, Yang Y, Cao J, Huang J, Chen J, Chen H, Zhang J, Wang L, Xu X, Gale RP, Wang L. Lei T, et al. Leukemia. 2024 Dec;38(12):2517-2543. doi: 10.1038/s41375-024-02444-y. Epub 2024 Oct 25. Leukemia. 2024. PMID: 39455854 Free PMC article. Review. - Contribution of CRISPRable DNA to human complex traits.
Zhai R, Zheng C, Yang Z, Li T, Chen J, Shen X. Zhai R, et al. Commun Biol. 2022 Oct 20;5(1):1111. doi: 10.1038/s42003-022-03969-7. Commun Biol. 2022. PMID: 36266475 Free PMC article. Review. - Detect-seq reveals out-of-protospacer editing and target-strand editing by cytosine base editors.
Lei Z, Meng H, Lv Z, Liu M, Zhao H, Wu H, Zhang X, Liu L, Zhuang Y, Yin K, Yan Y, Yi C. Lei Z, et al. Nat Methods. 2021 Jun;18(6):643-651. doi: 10.1038/s41592-021-01172-w. Epub 2021 Jun 7. Nat Methods. 2021. PMID: 34099937 - Gene Therapy in Rare Respiratory Diseases: What Have We Learned So Far?
Bañuls L, Pellicer D, Castillo S, Navarro-García MM, Magallón M, González C, Dasí F. Bañuls L, et al. J Clin Med. 2020 Aug 8;9(8):2577. doi: 10.3390/jcm9082577. J Clin Med. 2020. PMID: 32784514 Free PMC article. Review. - Brain-Targeted Cas12a Ribonucleoprotein Nanocapsules Enable Synergetic Gene Co-Editing Leading to Potent Inhibition of Orthotopic Glioblastoma.
Ruan W, Xu S, An Y, Cui Y, Liu Y, Wang Y, Ismail M, Liu Y, Zheng M. Ruan W, et al. Adv Sci (Weinh). 2024 Sep;11(33):e2402178. doi: 10.1002/advs.202402178. Epub 2024 Jun 28. Adv Sci (Weinh). 2024. PMID: 38943253 Free PMC article.
References
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials