Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair - PubMed (original) (raw)
Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Chenxu Zhu et al. Proc Natl Acad Sci U S A. 2016.
Abstract
NEIL1 (Nei-like 1) is a DNA repair glycosylase guarding the mammalian genome against oxidized DNA bases. As the first enzymes in the base-excision repair pathway, glycosylases must recognize the cognate substrates and catalyze their excision. Here we present crystal structures of human NEIL1 bound to a range of duplex DNA. Together with computational and biochemical analyses, our results suggest that NEIL1 promotes tautomerization of thymine glycol (Tg)-a preferred substrate-for optimal binding in its active site. Moreover, this tautomerization event also facilitates NEIL1-catalyzed Tg excision. To our knowledge, the present example represents the first documented case of enzyme-promoted tautomerization for efficient substrate recognition and catalysis in an enzyme-catalyzed reaction.
Keywords: QM/MM; base-excision repair; enzyme catalysis; glycosylase; substrate recognition.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
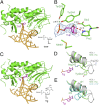
Fig. 1.
Crystal structures of NEIL1 bound to dsDNA containing THF and Tg, respectively. (A) Overall view of NEIL1 bound to a THF-containing duplex, with the flipped THF shown in purple. (B) Accommodation of THF in the active site of NEIL1. Density map (2_F_obs − _F_cal) of THF is shown. Hydrogen bonds are shown in green dashed lines. The red dashed line shows the distance between the α-amino group of Pro2 and C1 atom of THF. (C) Overall view of NEIL1 bound to a Tg-containing duplex, with the flipped Tg shown in purple. (D) Overlay of the apo and THF-bound NEIL1 structures, highlighting the conformational change of Arg242. In the Tg-bound structure, Arg242 in the flexible lesion recognition loop flips over, and the polar side chain of Arg242 points to the Tg base. Tyr244 resides approximately in the same location in both apo (gray) and Tg-bound (green) structures. (E) Overlay of the lesion recognition in the apo (gray), Tg (green)-, and THF (cyan)-bound structures. The same angle as in D is shown here for comparison.
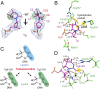
Fig. 2.
Tautomerization-dependent recognition of Tg. (A) Electron density map of Tg. The blue 2_F_obs − _F_cal map is contoured at 1.2σ and the green _F_obs − _F_cal omit map—by removing the 5-methyl (C7) and 6-hydroxyl group (O21) of Tg—is contoured at 3.0σ [omit maps removing 5-hydroxyl group (O22) are shown in SI Appendix, Fig. S3_A_]. (B) The active-site pocket of Tg-bound NEIL1. Hydrogen bonds are shown in green dashed lines. The hydrophobic pocket surrounding the 5-methyl group of Tg is indicated by a yellow curve. The N3 of Tg and Nη of Arg242 is highlighted with yellow background. (C) Tg tautomers in the lactam (Upper) and lactim (Lower) forms. (D) Optimized structure of the Tg-bound NEIL1 active site. Due to the Tg2-OH tautomer, a new hydrogen bond was observed between 2-OH of Tg and Glu6. Key distances are marked in black (in angstroms), and the 2-OH group of Tg is highlighted with yellow background.
Fig. 3.
Proposed mechanisms and computational verification. (A) Ribose-protonated pathway initiated with the NEIL1–R242–Tg2-OH structure. Along this pathway, we list the reactant (R2-1), two intermediate states (R2-2 and R2-3), and the product (R2-4). (B) The quantitative characterization of the above proposed mechanisms using QM/MM umbrella sampling (see SI Appendix, Detailed Materials and Methods, Section IX for more details): NEIL1–R242–Tg2-OH ribose-protonated pathway (full line), and NEIL1–R242–Tg4-OH ribose-protonated pathway (SI Appendix, Scheme S1) (dashed line). Numbers along the curves correspond to relative free energies of the transition states and intermediate states (including reactant and product).
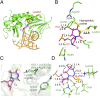
Fig. 4.
Structure of unedited NEIL1 (Lys242) bound to Tg. (A) Overall view of unedited NEIL1 bound to dsDNA containing Tg. Lys242 is highlighted in a red circle. (B) The active-site pocket of unedited NEIL1 bound to Tg. Hydrogen bonds are shown in green dashed lines. The N3 of Tg and Nζ of Lys242 is highlighted with yellow background. (C) Superposition of unedited (green) and edited (gray) NEIL1–Tg structures. The positions of the flipped base are almost identical, and both Arg242 and Lys242 point toward Tg. (D) Optimized structure of the Tg-bound NEIL1 (Lys242) active site. A hydrogen bond between 2-OH of Tg and Glu6 was also observed in this structure. Key distances are marked in black (in angstroms), and the 2-OH group is highlighted with yellow background.
Similar articles
- Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2.
Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Hailer MK, et al. DNA Repair (Amst). 2005 Jan 2;4(1):41-50. doi: 10.1016/j.dnarep.2004.07.006. DNA Repair (Amst). 2005. PMID: 15533836 - The Biochemical Role of the Human NEIL1 and NEIL3 DNA Glycosylases on Model DNA Replication Forks.
Albelazi MS, Martin PR, Mohammed S, Mutti L, Parsons JL, Elder RH. Albelazi MS, et al. Genes (Basel). 2019 Apr 23;10(4):315. doi: 10.3390/genes10040315. Genes (Basel). 2019. PMID: 31018584 Free PMC article. - Repair of thymine glycol by hNth1 and hNeil1 is modulated by base pairing and cis-trans epimerization.
Ocampo-Hafalla MT, Altamirano A, Basu AK, Chan MK, Ocampo JE, Cummings A Jr, Boorstein RJ, Cunningham RP, Teebor GW. Ocampo-Hafalla MT, et al. DNA Repair (Amst). 2006 Apr 8;5(4):444-54. doi: 10.1016/j.dnarep.2005.12.004. Epub 2006 Jan 30. DNA Repair (Amst). 2006. PMID: 16446124 - DNA glycosylases search for and remove oxidized DNA bases.
Wallace SS. Wallace SS. Environ Mol Mutagen. 2013 Dec;54(9):691-704. doi: 10.1002/em.21820. Epub 2013 Oct 7. Environ Mol Mutagen. 2013. PMID: 24123395 Free PMC article. Review. - New paradigms in the repair of oxidative damage in human genome: mechanisms ensuring repair of mutagenic base lesions during replication and involvement of accessory proteins.
Dutta A, Yang C, Sengupta S, Mitra S, Hegde ML. Dutta A, et al. Cell Mol Life Sci. 2015 May;72(9):1679-98. doi: 10.1007/s00018-014-1820-z. Epub 2015 Jan 10. Cell Mol Life Sci. 2015. PMID: 25575562 Free PMC article. Review.
Cited by
- Single-Molecule Titration in a Protein Nanoreactor Reveals the Protonation/Deprotonation Mechanism of a C:C Mismatch in DNA.
Ren H, Cheyne CG, Fleming AM, Burrows CJ, White HS. Ren H, et al. J Am Chem Soc. 2018 Apr 18;140(15):5153-5160. doi: 10.1021/jacs.8b00593. Epub 2018 Apr 3. J Am Chem Soc. 2018. PMID: 29562130 Free PMC article. - Heritable pattern of oxidized DNA base repair coincides with pre-targeting of repair complexes to open chromatin.
Bacolla A, Sengupta S, Ye Z, Yang C, Mitra J, De-Paula RB, Hegde ML, Ahmed Z, Mort M, Cooper DN, Mitra S, Tainer JA. Bacolla A, et al. Nucleic Acids Res. 2021 Jan 11;49(1):221-243. doi: 10.1093/nar/gkaa1120. Nucleic Acids Res. 2021. PMID: 33300026 Free PMC article. - Base excision repair of oxidative DNA damage: from mechanism to disease.
Whitaker AM, Schaich MA, Smith MR, Flynn TS, Freudenthal BD. Whitaker AM, et al. Front Biosci (Landmark Ed). 2017 Mar 1;22(9):1493-1522. doi: 10.2741/4555. Front Biosci (Landmark Ed). 2017. PMID: 28199214 Free PMC article. Review. - 2'-Fluorinated Hydantoins as Chemical Biology Tools for Base Excision Repair Glycosylases.
Cao S, Rogers J, Yeo J, Anderson-Steele B, Ashby J, David SS. Cao S, et al. ACS Chem Biol. 2020 Apr 17;15(4):915-924. doi: 10.1021/acschembio.9b00923. Epub 2020 Mar 13. ACS Chem Biol. 2020. PMID: 32069022 Free PMC article. - Dynamics of 5R-Tg Base Flipping in DNA Duplexes Based on Simulations─Agreement with Experiments and Beyond.
Wang SD, Eriksson LA, Zhang RB. Wang SD, et al. J Chem Inf Model. 2022 Jan 24;62(2):386-398. doi: 10.1021/acs.jcim.1c01169. Epub 2022 Jan 7. J Chem Inf Model. 2022. PMID: 34994562 Free PMC article.
References
- Dalhus B, Laerdahl JK, Backe PH, Bjørås M. DNA base repair--recognition and initiation of catalysis. FEMS Microbiol Rev. 2009;33(6):1044–1078. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous