Assembled Plastid and Mitochondrial Genomes, as well as Nuclear Genes, Place the Parasite Family Cynomoriaceae in the Saxifragales - PubMed (original) (raw)
Assembled Plastid and Mitochondrial Genomes, as well as Nuclear Genes, Place the Parasite Family Cynomoriaceae in the Saxifragales
Sidonie Bellot et al. Genome Biol Evol. 2016.
Abstract
Cynomoriaceae, one of the last unplaced families of flowering plants, comprise one or two species or subspecies of root parasites that occur from the Mediterranean to the Gobi Desert. Using Illumina sequencing, we assembled the mitochondrial and plastid genomes as well as some nuclear genes of a Cynomorium specimen from Italy. Selected genes were also obtained by Sanger sequencing from individuals collected in China and Iran, resulting in matrices of 33 mitochondrial, 6 nuclear, and 14 plastid genes and rDNAs enlarged to include a representative angiosperm taxon sampling based on data available in GenBank. We also compiled a new geographic map to discern possible discontinuities in the parasites' occurrence. Cynomorium has large genomes of 13.70-13.61 (Italy) to 13.95-13.76 pg (China). Its mitochondrial genome consists of up to 49 circular subgenomes and has an overall gene content similar to that of photosynthetic angiosperms, while its plastome retains only 27 of the normally 116 genes. Nuclear, plastid and mitochondrial phylogenies place Cynomoriaceae in Saxifragales, and we found evidence for several horizontal gene transfers from different hosts, as well as intracellular gene transfers.
Keywords: Cynomorium; Mediterranean-Irano-Turanian; chondriome; horizontal gene transfer; parasitic plants; plastome.
© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
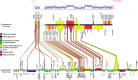
Fig. 1.—
Distribution and habit of_Cynomorium_. (A) Distribution range of_Cynomorium_obtained from the map in Hansen (1986) , relevant floras, and 203 GPS coordinates retrieved from the Global Biodiversity Information Facility (GBIF) portal in August 2015. Arrows indicate our own collections (details in table 1). Background map from Naturalearthdata.com. (B–J)_Cynomorium_plants from Italy (B, E, H), N. Cusimano and C. Cusimano 2, Iran (C and I)Zarre 59621, and China (D)_S.X. Luo 2014_from Tengger Desert; (F, G, J) G. Sun 1, from Gansu. Photos B to G by the respective collectors, photos H to J by N. Cusimano. (B–D) Plants in situ. (E) Part of a rhizome with young inflorescences and connected to the host roots (Atriplex portulacoides). (F) A fly (_Musca_spec.) visiting_Cynomorium_in the Tengger Desert. (G) Chinese plant connected to the host root (Nitraria tangutorum). (H) Young male and female flowers and a bract, showing a red stamen basis (arrow). (I) Stamen from herbarium material. (J) Male flowers showing white stamen bases (arrow). The color of the stamen basis has been used to differentiate_C. coccineum_subsp._songaricum_from_Cynomorium coccineum_subsp.coccineum.
Fig. 2.—
Gene losses and rearrangements in the plastome of_Cynomorium_compared to that of_Liquidambar formosana_(GenBank accession KC588388). Colored lines link homologous genes between the two plastomes. The LSC of_Cynomorium_is divided in two parts (linked by the dotted line) to help visualization. Maps were drawn using OGDraw ( Lohse et al. 2013 ). GC content and coverage of the_Cynomorium_plastome are depicted in blue and red, respectively. (The pattern is the same when using less stringent mapping parameters; Results.).
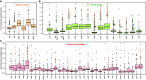
Fig. 3.—
Root to tip branch lengths of_Cynomorium_and other angiosperms in substitutions per site. Boxplots and open circles summarize the branch length distribution of: (A) nuclear, (B) plastid, and (C) mitochondrial genes of photosynthetic plants obtained from the constrained phylogenies in supplementary figures S3–S5 , Supplementary Material online. Black line: median; boxes: upper and lower quartile, including 50% of the data; whiskers: minimum and maximum of the data, provided that their length does not exceed 1.5× the interquartile range; open dots: outliers. Colored diamonds and circles represent, respectively, the branch length of the genes and of their copies found in other genomic compartments of_Cynomorium_: orange: gene copy located in the nuclear genome; green: gene copy located in the plastid genome; red: gene copy located in the mitochondrial genome; blue: gene copy amplified by Garcia et al. (2004) ; circles:_Cynomorium_gene copies amplified by Zhang et al. (2009) ; diamonds: all other_Cynomorium_sequences.
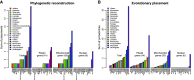
Fig. 4.—
Bar plots summarizing the analyses of single gene matrices when the rest of the topology was constrained to fit the currently accepted angiosperm relationships ( Angiosperm Phylogeny Group 2016 ). For simplicity, internally transferred gene copies were not included but their placements can be seen in supplementary figures S4–S6 , Supplementary Material online. (A) Results of ML tree searches. For each clade, the number of genes that placed_Cynomorium_sequence(s) in the respective clade is shown (details in supplementary figs. S4–S6 and table S4 , Supplementary Material online). (B) Results of evolutionary placement analyses (Materials and Methods). For each clade, the sum of the probabilities across all genes that placed_Cynomorium_in the respective clade is shown (details in supplementary fig. S7 , Supplementary Material online). For both analyses, results are shown overall and by genomic compartment. The asterisks indicate placements resulting exclusively from the sequences of Zhang et al. (2009) . Numbers of markers for each compartment are indicated in brackets.
Fig. 5.—
Phylogenetic trees obtained from ML analyses of the concatenated plastid (A) and nuclear (B) genes. Major angiosperm taxa are labeled following Angiosperm Phylogeny Group (2016) , with the orders including_Cynomorium_in purple (Saxifragales) and red (Fabids), and shown in more detail on the right.
Fig. 6.—
Phylogenetic tree obtained from ML analyses of the concatenated mitochondrial gene matrix. The genus label_Cynomorium_on the right refers to the placement of 26 genes from the Italian plant. The colored dots mark multiple copies of the respective gene; the two yellow diamonds mark a gene acquired by HGT, of which one native copy was included in the concatenated matrix; the single yellow square marks the_rps3_of a Chinese accession (see text).
Fig. 7.—
Neighbor net obtained from variable plastid regions of_Cynomorium_from Spain (Nickrent 4063), Italy (N. Cusimano and C. Cusimano 2), Iran (S. Zarre 59621), and China (L. Zhang 1 and S.X. Luo 618). The photos show habitats and hosts of_Cynomorium_at our collecting sites. The host of the Spanish sample is unknown. Photos by N. Cusimano (Italy and_Atriplex_), S.X. Luo (China and_Nitraria_), and A. Gröger (Iran and_Salsola_).
Similar articles
- Sequential horizontal gene transfers from different hosts in a widespread Eurasian parasitic plant, Cynomorium coccineum.
Cusimano N, Renner SS. Cusimano N, et al. Am J Bot. 2019 May;106(5):679-689. doi: 10.1002/ajb2.1286. Am J Bot. 2019. PMID: 31081928 - Degradation of key photosynthetic genes in the critically endangered semi-aquatic flowering plant Saniculiphyllum guangxiense (Saxifragaceae).
Folk RA, Sewnath N, Xiang CL, Sinn BT, Guralnick RP. Folk RA, et al. BMC Plant Biol. 2020 Jul 8;20(1):324. doi: 10.1186/s12870-020-02533-x. BMC Plant Biol. 2020. PMID: 32640989 Free PMC article. - Discovery of the photosynthetic relatives of the "Maltese mushroom" Cynomorium.
Nickrent DL, Der JP, Anderson FE. Nickrent DL, et al. BMC Evol Biol. 2005 Jun 21;5:38. doi: 10.1186/1471-2148-5-38. BMC Evol Biol. 2005. PMID: 15969755 Free PMC article. - Mitochondria in parasitic plants.
Petersen G, Anderson B, Braun HP, Meyer EH, Møller IM. Petersen G, et al. Mitochondrion. 2020 May;52:173-182. doi: 10.1016/j.mito.2020.03.008. Epub 2020 Mar 26. Mitochondrion. 2020. PMID: 32224234 Review. - The ins and outs of editing and splicing of plastid RNAs: lessons from parasitic plants.
Tillich M, Krause K. Tillich M, et al. N Biotechnol. 2010 Jul 31;27(3):256-66. doi: 10.1016/j.nbt.2010.02.020. Epub 2010 Mar 3. N Biotechnol. 2010. PMID: 20206308 Review.
Cited by
- Comparing complete organelle genomes of holoparasitic Christisonia kwangtungensis (Orabanchaceae) with its close relatives: how different are they?
Zhang C, Lin Q, Zhang J, Huang Z, Nan P, Li L, Song Z, Zhang W, Yang J, Wang Y. Zhang C, et al. BMC Plant Biol. 2022 Sep 17;22(1):444. doi: 10.1186/s12870-022-03814-3. BMC Plant Biol. 2022. PMID: 36114450 Free PMC article. - What we know so far and what we can expect next: A molecular investigation of plant parasitism.
Ishida JK, Costa EC. Ishida JK, et al. Genet Mol Biol. 2024 Sep 30;47Suppl 1(Suppl 1):e20240051. doi: 10.1590/1678-4685-GMB-2024-0051. eCollection 2024. Genet Mol Biol. 2024. PMID: 39348487 Free PMC article. - Complete chloroplast genome of the desert date (Balanites aegyptiaca (L.) Del. comparative analysis, and phylogenetic relationships among the members of Zygophyllaceae.
Al-Juhani WS, Alharbi SA, Al Aboud NM, Aljohani AY. Al-Juhani WS, et al. BMC Genomics. 2022 Aug 31;23(1):626. doi: 10.1186/s12864-022-08850-9. BMC Genomics. 2022. PMID: 36045328 Free PMC article. - Multichromosomal Mitochondrial Genome of Paphiopedilum micranthum: Compact and Fragmented Genome, and Rampant Intracellular Gene Transfer.
Yang JX, Dierckxsens N, Bai MZ, Guo YY. Yang JX, et al. Int J Mol Sci. 2023 Feb 16;24(4):3976. doi: 10.3390/ijms24043976. Int J Mol Sci. 2023. PMID: 36835385 Free PMC article. - Mitochondrial genome evolution in parasitic plants.
Zervas A, Petersen G, Seberg O. Zervas A, et al. BMC Evol Biol. 2019 Apr 8;19(1):87. doi: 10.1186/s12862-019-1401-8. BMC Evol Biol. 2019. PMID: 30961535 Free PMC article.
References
- Angiosperm Phylogeny Group . 2009. . An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III . Bot J Linn Soc . 161 : 105 – 121 .
- Angiosperm Phylogeny Group . 2016. . An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV . Bot J Linn Soc . 181 : 1 – 20 .
- Bellot S, Renner SS. 2014. . Exploring new dating approaches for parasites: the worldwide Apodanthaceae (Cucurbitales) as an example . Mol Phylogenet Evol. 80 : 1 – 10 . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases