Alignment-free microbial phylogenomics under scenarios of sequence divergence, genome rearrangement and lateral genetic transfer - PubMed (original) (raw)
Alignment-free microbial phylogenomics under scenarios of sequence divergence, genome rearrangement and lateral genetic transfer
Guillaume Bernard et al. Sci Rep. 2016.
Abstract
Alignment-free (AF) approaches have recently been highlighted as alternatives to methods based on multiple sequence alignment in phylogenetic inference. However, the sensitivity of AF methods to genome-scale evolutionary scenarios is little known. Here, using simulated microbial genome data we systematically assess the sensitivity of nine AF methods to three important evolutionary scenarios: sequence divergence, lateral genetic transfer (LGT) and genome rearrangement. Among these, AF methods are most sensitive to the extent of sequence divergence, less sensitive to low and moderate frequencies of LGT, and most robust against genome rearrangement. We describe the application of AF methods to three well-studied empirical genome datasets, and introduce a new application of the jackknife to assess node support. Our results demonstrate that AF phylogenomics is computationally scalable to multi-genome data and can generate biologically meaningful phylogenies and insights into microbial evolution.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
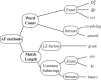
Figure 1. Alignment-free methods classification.
Classification of alignment-free methods, modified following Haubold. LZ: Lempel-Ziv.
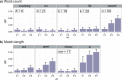
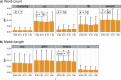
Figure 2. Accuracy of AF methods based on m.
RF distances are shown for (a) word-count methods and (b) match-length methods at m = 0.1, 0.5 and 0.9. Error bars indicate standard deviation from the mean across 50 replicates.
Figure 3. Accuracy of AF methods based on l.
RF distances are shown for (a) word-count methods and (b) match-length methods at l = 0, 5, 25, 125, 250 and 500. Error bars indicate standard deviation from the mean across 50 replicates.
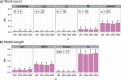
Figure 4. Accuracy of AF methods based on d.
RF distances are shown for (a) word-count methods and (b) match-length methods at d = 200, 1000, 3000 and 5000. Error bars indicate standard deviation from the mean across 50 replicates.
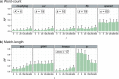
Figure 5. Accuracy of AF method based on r.
RF distances are shown for (a) word-count methods and (b) match-length methods at r = 0.00, 0.01, 0.10 and 1.00. Error bars indicate standard deviation from the mean across 50 replicates.
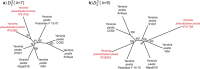
Figure 6. AF phylogeny of 143 prokaryote genomes.
Phylogenetic tree of 143 prokaryote genomes using  at k = 24, supported by JK values. The 15 phylum-level backbone nodes of Beiko et al. are marked with solid circles.
at k = 24, supported by JK values. The 15 phylum-level backbone nodes of Beiko et al. are marked with solid circles.
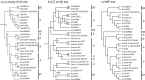
Figure 7. Phylogenetic trees of 27 E. coli and Shigella species.
(a) Tree generated using co-phylog at K = 8, supported by JK values. (b) Tree generated using  at k = 26, supported by JK values. (c) MRP tree constructed from 5282 Bayesian protein trees. Taxa labeled with an asterisk in each AF tree (a,b) are positioned differently in comparison to the reference (c). ECOR groups and Shigella (S) are indicated.
at k = 26, supported by JK values. (c) MRP tree constructed from 5282 Bayesian protein trees. Taxa labeled with an asterisk in each AF tree (a,b) are positioned differently in comparison to the reference (c). ECOR groups and Shigella (S) are indicated.
Figure 8. Phylogenetic trees of 8 Yersinia genomes.
(a) Tree generated using  at k = 7, supported by JK values. (b) Tree generated using
at k = 7, supported by JK values. (b) Tree generated using  at k = 9, supported by JK values.
at k = 9, supported by JK values.
Similar articles
- Inferring Phylogenomic Relationship of Microbes Using Scalable Alignment-Free Methods.
Bernard G, Stephens TG, González-Pech RA, Chan CX. Bernard G, et al. Methods Mol Biol. 2021;2242:69-76. doi: 10.1007/978-1-0716-1099-2_5. Methods Mol Biol. 2021. PMID: 33961218 - Next-generation phylogenomics.
Chan CX, Ragan MA. Chan CX, et al. Biol Direct. 2013 Jan 22;8:3. doi: 10.1186/1745-6150-8-3. Biol Direct. 2013. PMID: 23339707 Free PMC article. - Inferring phylogenies of evolving sequences without multiple sequence alignment.
Chan CX, Bernard G, Poirion O, Hogan JM, Ragan MA. Chan CX, et al. Sci Rep. 2014 Sep 30;4:6504. doi: 10.1038/srep06504. Sci Rep. 2014. PMID: 25266120 Free PMC article. - Phylogenomic networks.
Dagan T. Dagan T. Trends Microbiol. 2011 Oct;19(10):483-91. doi: 10.1016/j.tim.2011.07.001. Epub 2011 Aug 3. Trends Microbiol. 2011. PMID: 21820313 Review. - Detecting lateral genetic transfer : a phylogenetic approach.
Beiko RG, Ragan MA. Beiko RG, et al. Methods Mol Biol. 2008;452:457-69. doi: 10.1007/978-1-60327-159-2_21. Methods Mol Biol. 2008. PMID: 18566777 Review.
Cited by
- Inferring Phylogenomic Relationship of Microbes Using Scalable Alignment-Free Methods.
Bernard G, Stephens TG, González-Pech RA, Chan CX. Bernard G, et al. Methods Mol Biol. 2021;2242:69-76. doi: 10.1007/978-1-0716-1099-2_5. Methods Mol Biol. 2021. PMID: 33961218 - Evolutionary Insight into the Trypanosomatidae Using Alignment-Free Phylogenomics of the Kinetoplast.
Kaufer A, Stark D, Ellis J. Kaufer A, et al. Pathogens. 2019 Sep 18;8(3):157. doi: 10.3390/pathogens8030157. Pathogens. 2019. PMID: 31540520 Free PMC article. - Robust Inference of Genetic Exchange Communities from Microbial Genomes Using TF-IDF.
Cong Y, Chan YB, Phillips CA, Langston MA, Ragan MA. Cong Y, et al. Front Microbiol. 2017 Jan 19;8:21. doi: 10.3389/fmicb.2017.00021. eCollection 2017. Front Microbiol. 2017. PMID: 28154557 Free PMC article. - PhageGE: an interactive web platform for exploratory analysis and visualization of bacteriophage genomes.
Zhao J, Han J, Lin YW, Zhu Y, Aichem M, Garkov D, Bergen PJ, Nang SC, Ye JZ, Zhou T, Velkov T, Song J, Schreiber F, Li J. Zhao J, et al. Gigascience. 2024 Jan 2;13:giae074. doi: 10.1093/gigascience/giae074. Gigascience. 2024. PMID: 39320317 Free PMC article. - Synonymous nucleotide changes drive papillomavirus evolution.
King KM, Rajadhyaksha EV, Tobey IG, Van Doorslaer K. King KM, et al. Tumour Virus Res. 2022 Dec;14:200248. doi: 10.1016/j.tvr.2022.200248. Epub 2022 Oct 17. Tumour Virus Res. 2022. PMID: 36265836 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources