The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins - PubMed (original) (raw)
The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins
Andrew D Rouillard et al. Database (Oxford). 2016.
Abstract
Genomics, epigenomics, transcriptomics, proteomics and metabolomics efforts rapidly generate a plethora of data on the activity and levels of biomolecules within mammalian cells. At the same time, curation projects that organize knowledge from the biomedical literature into online databases are expanding. Hence, there is a wealth of information about genes, proteins and their associations, with an urgent need for data integration to achieve better knowledge extraction and data reuse. For this purpose, we developed the Harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins from over 70 major online resources. We extracted, abstracted and organized data into ∼72 million functional associations between genes/proteins and their attributes. Such attributes could be physical relationships with other biomolecules, expression in cell lines and tissues, genetic associations with knockout mouse or human phenotypes, or changes in expression after drug treatment. We stored these associations in a relational database along with rich metadata for the genes/proteins, their attributes and the original resources. The freely available Harmonizome web portal provides a graphical user interface, a web service and a mobile app for querying, browsing and downloading all of the collected data. To demonstrate the utility of the Harmonizome, we computed and visualized gene-gene and attribute-attribute similarity networks, and through unsupervised clustering, identified many unexpected relationships by combining pairs of datasets such as the association between kinase perturbations and disease signatures. We also applied supervised machine learning methods to predict novel substrates for kinases, endogenous ligands for G-protein coupled receptors, mouse phenotypes for knockout genes, and classified unannotated transmembrane proteins for likelihood of being ion channels. The Harmonizome is a comprehensive resource of knowledge about genes and proteins, and as such, it enables researchers to discover novel relationships between biological entities, as well as form novel data-driven hypotheses for experimental validation.Database URL: http://amp.pharm.mssm.edu/Harmonizome.
© The Author(s) 2016. Published by Oxford University Press.
Figures
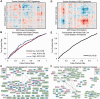
Figure 1.
Hierarchical clustering of gene-term, term-term and gene-gene matrices. (A) Gene-phenotype associations from the MPO organized into a binary matrix and clustered using hierarchical clustering. (B) Zooming into a cluster of genes with similar associated phenotypes, filtered to show higher level phenotypes associated with at least half of the genes in the cluster but no > 10% of all genes. (C) The gene–gene and cell-line/cell-line similarity matrices are from the CCLE gene expression dataset. Along the main diagonal of both matrices, there are several distinct zones of high red intensity, indicating clusters of cell lines with similar differentially expressed genes (DEGs) and clusters of genes with similar patterns of expression across cell lines. (D) Zooming into the lung cancer cell-lines cluster.
Figure 2.
Example of combining datasets: matching kinases with diseases and drugs. (A) Hierarchical clustering of kinase perturbation signatures extracted from GEO and disease signatures extracted from GEO. (B) Validation of kinase-disease associations with genomics datasets. ROC curve showing concordance of kinase-disease associations derived by comparing gene expression profiles and kinase-disease associations collected from GWAS and other genetic association datasets. Low, medium and high labels correspond to confidence levels of associations from GWAS datasets. (C) Network showing top predictions of drug-kinase-disease associations. Red edges indicate kinase-disease associations that have supporting GWAS evidence. (D) Hierarchical clustering of signatures of DEGs for kinase perturbations extracted from GEO compared with signatures for cancer cell lines from CCLE. (E) ROC curve showing concordance of kinase-cell line associations derived by comparing gene expression profiles and driver kinase mutations for cell lines from COSMIC. (F) Network showing top predictions of drug-kinase-cell line associations. Red edges indicate kinase-cell line associations supported by COSMIC as having a driver mutation in the cell line.
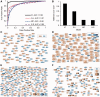
Figure 3.
Example of supervised machine learning: classifiers to predict ion channels (IC), phenotypes of single gene knockouts in mice (MP), ligands of GPCRs (G-L), and substrates of kinases (K-S). (A) ROC curve of the classifiers. (B) MCC as a function of the fraction of correct predictions. (C) Network showing candidate ion channels, predicted at a false discovery rate (FDR) of 0.67, connected to their most similar known ion channels, and limited to no more than three edges per node. (D) Network showing candidate gene-phenotype associations, predicted at a FDR of 0.33, limited to no more than three edges per node, and trimmed to remove clusters with all edges supported by prior knowledge. Red edges indicate known associations. (E) Network showing candidate GPCR-ligand interactions; predicted at a FDR of 0.67 and limited to no more than three edges per node. Red edges indicate known interactions. (F) Network showing candidate kinase-substrate interactions predicted at a FDR of 0.67 and limited to no more than three edges per node. Red edges indicate known interactions.
Similar articles
- miRiaD: A Text Mining Tool for Detecting Associations of microRNAs with Diseases.
Gupta S, Ross KE, Tudor CO, Wu CH, Schmidt CJ, Vijay-Shanker K. Gupta S, et al. J Biomed Semantics. 2016 Apr 29;7(1):9. doi: 10.1186/s13326-015-0044-y. J Biomed Semantics. 2016. PMID: 27216254 Free PMC article. - Getting Started with the IDG KMC Datasets and Tools.
Kropiwnicki E, Binder JL, Yang JJ, Holmes J, Lachmann A, Clarke DJB, Sheils T, Kelleher KJ, Metzger VT, Bologa CG, Oprea TI, Ma'ayan A. Kropiwnicki E, et al. Curr Protoc. 2022 Jan;2(1):e355. doi: 10.1002/cpz1.355. Curr Protoc. 2022. PMID: 35085427 Free PMC article. - LiverCancerMarkerRIF: a liver cancer biomarker interactive curation system combining text mining and expert annotations.
Dai HJ, Wu JC, Lin WS, Reyes AJ, Dela Rosa MA, Syed-Abdul S, Tsai RT, Hsu WL. Dai HJ, et al. Database (Oxford). 2014 Aug 27;2014:bau085. doi: 10.1093/database/bau085. Print 2014. Database (Oxford). 2014. PMID: 25168057 Free PMC article. - Combining literature text mining with microarray data: advances for system biology modeling.
Faro A, Giordano D, Spampinato C. Faro A, et al. Brief Bioinform. 2012 Jan;13(1):61-82. doi: 10.1093/bib/bbr018. Epub 2011 Jun 15. Brief Bioinform. 2012. PMID: 21677032 Review. - Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.
Wolahan SM, Hirt D, Glenn TC. Wolahan SM, et al. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review.
Cited by
- Non-electrophilic NRF2 activators promote wound healing in human keratinocytes and diabetic mice and demonstrate selective downstream gene targeting.
Barakat M, Han C, Chen L, David BP, Shi J, Xu A, Skowron KJ, Johnson T, Woods RA, Ankireddy A, Reddy SP, Moore TW, DiPietro LA. Barakat M, et al. Sci Rep. 2024 Oct 24;14(1):25258. doi: 10.1038/s41598-024-75786-3. Sci Rep. 2024. PMID: 39448644 Free PMC article. - Selective autophagy of AKAP11 activates cAMP/PKA to fuel mitochondrial metabolism and tumor cell growth.
Deng Z, Li X, Blanca Ramirez M, Purtell K, Choi I, Lu JH, Yu Q, Yue Z. Deng Z, et al. Proc Natl Acad Sci U S A. 2021 Apr 6;118(14):e2020215118. doi: 10.1073/pnas.2020215118. Proc Natl Acad Sci U S A. 2021. PMID: 33785595 Free PMC article. - Cocaine-related DNA methylation in caudate neurons alters 3D chromatin structure of the IRXA gene cluster.
Vaillancourt K, Yang J, Chen GG, Yerko V, Théroux JF, Aouabed Z, Lopez A, Thibeault KC, Calipari ES, Labonté B, Mechawar N, Ernst C, Nagy C, Forné T, Nestler EJ, Mash DC, Turecki G. Vaillancourt K, et al. Mol Psychiatry. 2021 Jul;26(7):3134-3151. doi: 10.1038/s41380-020-00909-x. Epub 2020 Oct 12. Mol Psychiatry. 2021. PMID: 33046833 Free PMC article. - Elucidating prognosis in cervical squamous cell carcinoma and endocervical adenocarcinoma: a novel anoikis-related gene signature model.
Wang M, Ying Q, Ding R, Xing Y, Wang J, Pan Y, Pan B, Xiang G, Liu Z. Wang M, et al. Front Oncol. 2024 Jun 26;14:1352638. doi: 10.3389/fonc.2024.1352638. eCollection 2024. Front Oncol. 2024. PMID: 38988712 Free PMC article. - Comparative analysis identifies genetic and molecular factors associated with prognostic clusters of PANoptosis in glioma, kidney and melanoma cancer.
Mall R, Kanneganti TD. Mall R, et al. Sci Rep. 2023 Nov 28;13(1):20962. doi: 10.1038/s41598-023-48098-1. Sci Rep. 2023. PMID: 38017056 Free PMC article.
References
MeSH terms
Grants and funding
- R01 GM098316/GM/NIGMS NIH HHS/United States
- T32 HL007824/HL/NHLBI NIH HHS/United States
- U54 CA189201/CA/NCI NIH HHS/United States
- U54 HL127624/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources