Polymer physics of chromosome large-scale 3D organisation - PubMed (original) (raw)
Polymer physics of chromosome large-scale 3D organisation
Andrea M Chiariello et al. Sci Rep. 2016.
Abstract
Chromosomes have a complex architecture in the cell nucleus, which serves vital functional purposes, yet its structure and folding mechanisms remain still incompletely understood. Here we show that genome-wide chromatin architecture data, as mapped by Hi-C methods across mammalian cell types and chromosomes, are well described by classical scaling concepts of polymer physics, from the sub-Mb to chromosomal scales. Chromatin is a complex mixture of different regions, folded in the conformational classes predicted by polymer thermodynamics. The contact matrix of the Sox9 locus, a region linked to severe human congenital diseases, is derived with high accuracy in mESCs and its molecular determinants identified by the theory; Sox9 self-assembles hierarchically in higher-order domains, involving abundant many-body contacts. Our approach is also applied to the Bmp7 locus. Finally, the model predictions on the effects of mutations on folding are tested against available data on a deletion in the Xist locus. Our results can help progressing new diagnostic tools for diseases linked to chromatin misfolding.
Figures
Figure 1. A polymer model of chromatin.
(a) The Strings&Binders (SBS) model is a Self-Avoiding (SAW) chain of beads interacting with molecular binders having a concentration, c, and a binding affinity E Int. (b) Polymer physics dictates that the model different stable architectural classes correspond to the different phases of its phase diagram: at low E Int or c, the polymer is open and randomly folded in its coil phase; above its Θ-point transition, in the globule phase it is closed in more compact conformations as signalled by a drop in its gyration radius (Figure S1a); in the closed state, at higher values of E Int or c, its binders have a transition form a disordered to an ordered arrangement, as shown by their Structure Factor (Figure S1b).
Figure 2. Chromatin is a mixture of regions folded in different thermodynamics states.
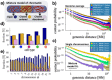
(a) We model a chromatin filament as a mixture of differently folded regions, each belonging to one of the stable conformational classes envisaged by polymer physics (Fig. 1). In this view, the average pairwise contact probability is only determined by the relative abundances of the states in the mixture, as each state has a fixed, specific pairwise contact probability (Figure S2). (b) Genome-wide average contact frequencies across human cell types, from Hi-C and TCC technologies, can be fitted from the sub-Mb to chromosomal scales by such a mixture model. (c) Single chromosome data (here from IMR90 cells) can be similarly explained. (d) Different cell types (see colour scheme in panel b) have a different chromatin composition, with hESC (orange circle) more open than differentiated cells, such as IMR90 (blue circle). (e) Within a given cell type (here IMR90, as in c) distinct chromosomes have also a different composition, with chromosome X formed mostly of closed regions, whereas gene rich chromosomes, e.g., chr.19, are up to 70% open.
Figure 3. Higher-order domains and symmetry breaking architectures.
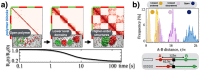
(a) We consider a block-copolymer composed of red and green binding domains which spontaneously folds, from an initially SAW conformation, in the ordered closed state of Fig. 1 (here c = 54 n_mol/l_ and E int = 4.1 kBT). The process is marked by a decrease of the gyration radius, R g(t), in time (bottom panel) and by the formation of a hierarchy of higher-order domains, as reflected by the contact matrix pattern (top). (b) Pairs of sites with the same contour separation, differently positioned across a block boundary (see bottom panel), have the same average physical distances, r, in the open phase. Yet, in the closed states, the symmetry is broken by their different position relative to the boundary as the two pairs have a different physical distance, as seen from the corresponding distributions of r (from left to right E int = 4.1, 3.1, 0 kBT, and c = 54 n_mol/l_).
Figure 4. Polymer physics captures the folding of the Sox9 locus.
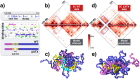
(a) Top: the considered Sox9 locus in mESC-J1 cells, with a few marker genes. Bottom: the SBS polymer model that best explain the Hi-C contact map of the Sox9 region has the shown different types of binding sites, as seen in the zoom (different colors); their abundance is represented as an histogram over the genomic sequence. The bar at the bottom highlights three main regional areas to help 3D visualization. (b) The model derived pairwise contact frequency matrix (bottom) has a 95% Pearson correlation with Hi-C experimental data (top). (c) A snapshot of the Sox9 locus in its closed disordered state as derived by the polymer model, with the position of TSSs of some key genes highlighted. (d) In the locus, many-body contacts of n sites are exponentially more abundant than in random SAW conformations (the ratio of the average number is plotted v.s. n), which could help the simultaneous co-localization of multiple functional regulatory regions. (e) The Sox9 locus folding dynamics from an initially open conformation towards the closed disordered state is represented by the gyration radius, R g(t). Chromatin domains self-assemble hierarchically in higher-order structures, in approx. 20_s_.
Figure 5. Model predicted effects of the Δ_XTX_ deletion in the Xist locus.
(a) The different binding domains are shown of the SBS polymer model of the Xist locus, inferred from 5C contact data in mESC-E14 cells. To help 3D visualization, the bars at the bottom highlight in cyan the wild-type region that is deleted in Δ_XTX_ cells, its flanking sequences in yellow and the remaining regional areas overlapping the locus two TADs in violet and blue. (b) The model inferred contact matrix (bottom) has a 96% Pearson correlation with 5C experimental data (top). (c) A snapshot of the Xist locus in its closed disordered state. (d) The contact matrix predicted, under no adjustable parameters, by the very same model after the Δ_XTX_ deletion (bottom) has a pattern of ectopic interactions (full line magenta box) w.r.t. wild-type data (panel b) very similar to the one seen in the corresponding experimental 5C data (top, correlation 91%). (e) Ectopic interactions extend across the yellow regions flanking the Δ_XTX_ deletion, as visible in the snapshot of the deleted locus (closed disordered state).
Similar articles
- Predicting chromatin architecture from models of polymer physics.
Bianco S, Chiariello AM, Annunziatella C, Esposito A, Nicodemi M. Bianco S, et al. Chromosome Res. 2017 Mar;25(1):25-34. doi: 10.1007/s10577-016-9545-5. Epub 2017 Jan 9. Chromosome Res. 2017. PMID: 28070687 Review. - De novo prediction of human chromosome structures: Epigenetic marking patterns encode genome architecture.
Di Pierro M, Cheng RR, Lieberman Aiden E, Wolynes PG, Onuchic JN. Di Pierro M, et al. Proc Natl Acad Sci U S A. 2017 Nov 14;114(46):12126-12131. doi: 10.1073/pnas.1714980114. Epub 2017 Oct 31. Proc Natl Acad Sci U S A. 2017. PMID: 29087948 Free PMC article. - Physics-Based Polymer Models to Probe Chromosome Structure in Single Molecules.
Conte M, Chiariello AM, Bianco S, Esposito A, Abraham A, Nicodemi M. Conte M, et al. Methods Mol Biol. 2023;2655:57-66. doi: 10.1007/978-1-0716-3143-0_5. Methods Mol Biol. 2023. PMID: 37212988 - Polymer models of the hierarchical folding of the Hox-B chromosomal locus.
Annunziatella C, Chiariello AM, Bianco S, Nicodemi M. Annunziatella C, et al. Phys Rev E. 2016 Oct;94(4-1):042402. doi: 10.1103/PhysRevE.94.042402. Epub 2016 Oct 4. Phys Rev E. 2016. PMID: 27841585 - Molecular Dynamics simulations of the Strings and Binders Switch model of chromatin.
Annunziatella C, Chiariello AM, Esposito A, Bianco S, Fiorillo L, Nicodemi M. Annunziatella C, et al. Methods. 2018 Jun 1;142:81-88. doi: 10.1016/j.ymeth.2018.02.024. Epub 2018 Mar 6. Methods. 2018. PMID: 29522804 Review.
Cited by
- Polymer Physics Models Reveal Structural Folding Features of Single-Molecule Gene Chromatin Conformations.
Conte M, Abraham A, Esposito A, Yang L, Gibcus JH, Parsi KM, Vercellone F, Fontana A, Di Pierno F, Dekker J, Nicodemi M. Conte M, et al. Int J Mol Sci. 2024 Sep 23;25(18):10215. doi: 10.3390/ijms251810215. Int J Mol Sci. 2024. PMID: 39337699 Free PMC article. - RNA-mediated symmetry breaking enables singular olfactory receptor choice.
Pourmorady AD, Bashkirova EV, Chiariello AM, Belagzhal H, Kodra A, Duffié R, Kahiapo J, Monahan K, Pulupa J, Schieren I, Osterhoudt A, Dekker J, Nicodemi M, Lomvardas S. Pourmorady AD, et al. Nature. 2024 Jan;625(7993):181-188. doi: 10.1038/s41586-023-06845-4. Epub 2023 Dec 20. Nature. 2024. PMID: 38123679 Free PMC article. - Evaluating the role of the nuclear microenvironment in gene function by population-based modeling.
Yildirim A, Hua N, Boninsegna L, Zhan Y, Polles G, Gong K, Hao S, Li W, Zhou XJ, Alber F. Yildirim A, et al. Nat Struct Mol Biol. 2023 Aug;30(8):1193-1206. doi: 10.1038/s41594-023-01036-1. Epub 2023 Aug 14. Nat Struct Mol Biol. 2023. PMID: 37580627 Free PMC article. - Polymer physics predicts the effects of structural variants on chromatin architecture.
Bianco S, Lupiáñez DG, Chiariello AM, Annunziatella C, Kraft K, Schöpflin R, Wittler L, Andrey G, Vingron M, Pombo A, Mundlos S, Nicodemi M. Bianco S, et al. Nat Genet. 2018 May;50(5):662-667. doi: 10.1038/s41588-018-0098-8. Epub 2018 Apr 16. Nat Genet. 2018. PMID: 29662163 - A Lamin-Associated Chromatin Model for Chromosome Organization.
Maji A, Ahmed JA, Roy S, Chakrabarti B, Mitra MK. Maji A, et al. Biophys J. 2020 Jun 16;118(12):3041-3050. doi: 10.1016/j.bpj.2020.05.014. Epub 2020 May 20. Biophys J. 2020. PMID: 32492372 Free PMC article.
References
- Misteli T. Beyond the sequence: cellular organisation of genome function. Cell 128, 787–800 (2007). - PubMed
- Tanay A. & Cavalli G. Chromosomal domains: epigenetic contexts and functional implications of genomic compartmentalization. Curr. Op. Gen. & Dev. 23, 197–203 (2013). - PubMed
- Bickmore W. A. & van Steensel B. Genome architecture: domain organisation of interphase chromosomes. Cell 152, 1270–84 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous