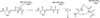SkfB Abstracts a Hydrogen Atom from Cα on SkfA To Initiate Thioether Cross-Link Formation - PubMed (original) (raw)
SkfB Abstracts a Hydrogen Atom from Cα on SkfA To Initiate Thioether Cross-Link Formation
Nathan A Bruender et al. Biochemistry. 2016.
Abstract
Sulfur to α-carbon thioether-containing peptides (sactipeptides) are ribosomally synthesized post-translationally modified peptides with bacteriocidal activities. The thioether cross-link, which is required for biological activity, is installed by a member of the radical S-adenosyl-l-methionine (SAM) superfamily in the peptide substrate. Herein, we show that the radical SAM enzyme, SkfB, utilizes the 5'-deoxyadenosyl radical generated from the reductive cleavage of SAM to abstract a hydrogen atom from the α-carbon of the amino acid at position 12 in the substrate, SkfA, to initiate the installation of a thioether cross-link. The insights from this work can be applied to all radical SAM sactipeptide maturases.
Figures
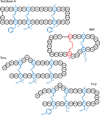
Figure 1
Schematic of the sactipeptides subtilosin A, sporulation killing factor (SKF), and the two peptides, Trnα and Trnβ. The sulfur to α-carbon thioether crosslinks are highlighted in blue. The sidechains of the accepting residues remain unchanged.
Figure 2
Mass spectra of (A) dAdo or (B and C) SkfA isolated from reactions containing (a) wild-type, (b) M12[β-D3]A–, or (c) M12[U-D4]A–SkfA. (A) The mass spectra show the monoisotopic (m/z = 252.1079 – 252.1083) and the corresponding 15N (m/z = 253.1049 – 253.1056), 13C (m/z = 253.1112 – 253.1126), and 2H isotope peaks (m/z = 253.1138 – 253.1146) of dAdo. (B) Mass spectra of SkfA peptides zooming in on the +5 charge state (see Fig S4a–c for the full mass spectra). The peak indicated by • corresponds to SkfA with three carbamidomethylated cysteines, whereas the * corresponds to SkfA containing one thioether crosslink and two carbamidomethylated cysteines. (C) The deconvoluted mass spectra generated using the Xtract software (Thermo Fisher) from the full mass spectra shown in Fig S4a–c.
Scheme 1
A proposed mechanism of thioether crosslink formation in sactipeptides. The dAdo• abstracts the Cα hydrogen atom from the acceptor residue to initiate catalysis to generate the captodatively stabilized radical intermediate.
Similar articles
- A Radical Clock Probe Uncouples H Atom Abstraction from Thioether Cross-Link Formation by the Radical S-Adenosyl-l-methionine Enzyme SkfB.
Kincannon WM, Bruender NA, Bandarian V. Kincannon WM, et al. Biochemistry. 2018 Aug 14;57(32):4816-4823. doi: 10.1021/acs.biochem.8b00537. Epub 2018 Jul 24. Biochemistry. 2018. PMID: 29965747 Free PMC article. - Two [4Fe-4S] clusters containing radical SAM enzyme SkfB catalyze thioether bond formation during the maturation of the sporulation killing factor.
Flühe L, Burghaus O, Wieckowski BM, Giessen TW, Linne U, Marahiel MA. Flühe L, et al. J Am Chem Soc. 2013 Jan 23;135(3):959-62. doi: 10.1021/ja310542g. Epub 2013 Jan 9. J Am Chem Soc. 2013. PMID: 23282011 - Bioinformatic Mapping of Radical S-Adenosylmethionine-Dependent Ribosomally Synthesized and Post-Translationally Modified Peptides Identifies New Cα, Cβ, and Cγ-Linked Thioether-Containing Peptides.
Hudson GA, Burkhart BJ, DiCaprio AJ, Schwalen CJ, Kille B, Pogorelov TV, Mitchell DA. Hudson GA, et al. J Am Chem Soc. 2019 May 22;141(20):8228-8238. doi: 10.1021/jacs.9b01519. Epub 2019 May 13. J Am Chem Soc. 2019. PMID: 31059252 Free PMC article. - The biosynthesis of thiol- and thioether-containing cofactors and secondary metabolites catalyzed by radical S-adenosylmethionine enzymes.
Jarrett JT. Jarrett JT. J Biol Chem. 2015 Feb 13;290(7):3972-9. doi: 10.1074/jbc.R114.599308. Epub 2014 Dec 4. J Biol Chem. 2015. PMID: 25477512 Free PMC article. Review. - Radical S-adenosylmethionine enzyme catalyzed thioether bond formation in sactipeptide biosynthesis.
Flühe L, Marahiel MA. Flühe L, et al. Curr Opin Chem Biol. 2013 Aug;17(4):605-12. doi: 10.1016/j.cbpa.2013.06.031. Epub 2013 Jul 24. Curr Opin Chem Biol. 2013. PMID: 23891473 Review.
Cited by
- Direct Detection of the α-Carbon Radical Intermediate Formed by OspD: Mechanistic Insights into Radical _S_-Adenosyl-l-methionine Peptide Epimerization.
Walls WG, Vagstad AL, Delridge T, Piel J, Broderick WE, Broderick JB. Walls WG, et al. J Am Chem Soc. 2024 Feb 28;146(8):5550-5559. doi: 10.1021/jacs.3c13829. Epub 2024 Feb 16. J Am Chem Soc. 2024. PMID: 38364824 Free PMC article. - Hydrogen-Deuterium Exchange Mass Spectrometry Identifies Local and Long-Distance Interactions within the Multicomponent Radical SAM Enzyme, PqqE.
Zhu W, Iavarone AT, Klinman JP. Zhu W, et al. ACS Cent Sci. 2024 Jan 17;10(2):251-263. doi: 10.1021/acscentsci.3c01023. eCollection 2024 Feb 28. ACS Cent Sci. 2024. PMID: 38435514 Free PMC article. - How a Subfamily of Radical S-Adenosylmethionine Enzymes Became a Mainstay of Ribosomally Synthesized and Post-translationally Modified Peptide Discovery.
Mendauletova A, Kostenko A, Lien Y, Latham J. Mendauletova A, et al. ACS Bio Med Chem Au. 2021 Dec 2;2(1):53-59. doi: 10.1021/acsbiomedchemau.1c00045. eCollection 2022 Feb 16. ACS Bio Med Chem Au. 2021. PMID: 37102180 Free PMC article. Review. - Sactipeptide Engineering by Probing the Substrate Tolerance of a Thioether-Bond-Forming Sactisynthase.
Ali A, Happel D, Habermann J, Schoenfeld K, Macarrón Palacios A, Bitsch S, Englert S, Schneider H, Avrutina O, Fabritz S, Kolmar H. Ali A, et al. Angew Chem Int Ed Engl. 2022 Nov 7;61(45):e202210883. doi: 10.1002/anie.202210883. Epub 2022 Oct 12. Angew Chem Int Ed Engl. 2022. PMID: 36049110 Free PMC article. - Biosynthesis of the sactipeptide Ruminococcin C by the human microbiome: Mechanistic insights into thioether bond formation by radical SAM enzymes.
Balty C, Guillot A, Fradale L, Brewee C, Lefranc B, Herrero C, Sandström C, Leprince J, Berteau O, Benjdia A. Balty C, et al. J Biol Chem. 2020 Dec 4;295(49):16665-16677. doi: 10.1074/jbc.RA120.015371. Epub 2020 Sep 24. J Biol Chem. 2020. PMID: 32972973 Free PMC article.
References
- Finking R, Marahiel MA. Ann Rev Microbiol. 2004;58:453–488. - PubMed
- Fischbach MA, Walsh CT. Chem Rev. 2006;106:3468–3496. - PubMed
- Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJ, Rebuffat S, Ross RP, Sahl HG, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Sussmuth RD, Tagg JR, Tang GL, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. Nat Prod Rep. 2013;30:108–160. - PMC - PubMed
- Babasaki K, Takao T, Shimonishi Y, Kurahashi K. J Biochemistry. 1985;98:585–603. - PubMed
- Gonzalez-Pastor JE. Science. 2003;301:510–513. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases