Decoding molecular interactions in microbial communities - PubMed (original) (raw)
Review
Decoding molecular interactions in microbial communities
Nicole A Abreu et al. FEMS Microbiol Rev. 2016 Sep.
Abstract
Microbial communities govern numerous fundamental processes on earth. Discovering and tracking molecular interactions among microbes is critical for understanding how single species and complex communities impact their associated host or natural environment. While recent technological developments in DNA sequencing and functional imaging have led to new and deeper levels of understanding, we are limited now by our inability to predict and interpret the intricate relationships and interspecies dependencies within these communities. In this review, we highlight the multifaceted approaches investigators have taken within their areas of research to decode interspecies molecular interactions that occur between microbes. Understanding these principles can give us greater insight into ecological interactions in natural environments and within synthetic consortia.
Keywords: corrinoids; metagenomics; microbial community; microbiome; molecular interactions; synthetic biology.
© FEMS 2016. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
Graphical Abstract Figure.
Understanding how microbes interact is key to deciphering microbial community assembly and stability; approaches that span whole communities to single isolate analyses, as well as sequence-function and synthetic approaches, lead to a deeper understanding of microbial interactions.
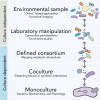
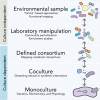
Figure 1.
Integrated, multilayered approaches needed to decode microbiomes. Whole communities can be studied by culture-independent approaches performed directly on environmental samples or on communities cultivated in the laboratory. Culture-dependent approaches, in which defined consortia, cocultures or single microbes are examined by a variety of methods in the laboratory, enable more detailed studies about molecular interactions in a subset of the community.
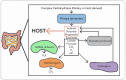
Figure 2.
Simplified model of carbon flow in the human gut. Complex carbohydrates obtained from the diet or host mucin are broken down by saccharolytic primary fermenters, usually Bacteroides and Bifidobacterium spp (Willis et al. 1996). Some end products of this fermentation (acetate, butyrate and propionate) are absorbed by the host. Other products such as glucose (not shown) and lactate are fermented to butyrate by bacteria such as Clostridium, Eubacterium and Fusobacterium spp. (Bourriaud et al. ; Belenguer et al. 2006). Consumption of formate, CO2 and H2 by acetogens (like Blautia hydrogenotrophica and Marvinbryantia formatexigens) prevents their buildup and provides additional acetate. Methanogens (such as M. smithii) and SRBs (such as D. piger), which are present in 30% and 50% of individuals (Hansen et al. ; Rey et al. 2013), also consume fermentation products to generate methane and hydrogen sulfide, respectively (Loubinoux et al. ; Samuel and Gordon 2006).
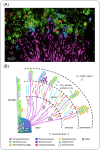
Figure 3.
Localization of oral microbial taxa in dental plaque. (A) CLASI-FISH imaging of plaque microbial communities. (B) Schematic of the spatial distribution of bacteria based on CLASI-FISH images. (Reprinted with permission from Mark Welch et al. .)
Figure 4.
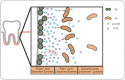
Model of A. actinomycetemcomitans (Aa) and S. gordonii (Sg) interaction in a mouse infection model. Sg adheres to the tooth pellicle surface and produces lactate, the preferred carbon source for Aa. Sg also produces hydrogen peroxide, which kills Aa at higher concentrations and induces production of biofilm dispersant by Aa (‘Flight’ zone). In the ‘Fight’ zone, catalase secreted by Aa detoxifies hydrogen peroxide effectively, and dispersant production is downregulated, allowing biofilm formation. In this zone, Aa has access to lactate. In the starvation zone, the lactate concentration is insufficient to promote growth of Aa (Stacy et al. 2014).
Figure 5.
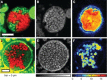
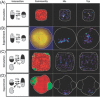
Imaging and metabolic profiling of ANME/SRB aggregates treated with 15N2. (A–C) ANME/SRB consortium with sulfate. (D–F) ANME/SRB consortium with AQDS as the sole electron acceptor. (A and D) FISH images highlighting AMNE in red and Desulfobacteriaceae in green. (B and E) NanoSIMS detection of 12C14N, indicating total cellular biomass. (C and F) NanoSIMS detection of 15N. (Reprinted with permission from Scheller et al. .)
Figure 6.
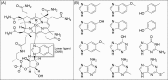
Corrinoid structure. (A) The structure of cobalamin is shown with the lower ligand, DMB, boxed. R represents the upper ligand (see Box 3). (B) Structures of commonly detected lower ligand bases.
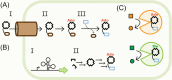
Figure 7.
Decoding corrinoid specificity. (A) In corrinoid-requiring organisms, specificity for particular corrinoid structures can be encoded in (I) corrinoid transporters, (II) corrinoid adenosyltransferases (Ado represents 5′-deoxyadenosine) and (III) corrinoid remodeling enzymes. (B) In corrinoid-producing organisms, specificity can be encoded in (I) the regulation of corrinoid-related genes (e.g. in corrinoid-binding riboswitches) and (II) the corrinoid biosynthetic pathway. (C) Corrinoid-dependent enzymes may also be specific for particular corrinoids.
Figure 8.
Theoretical model and experimental evidence for spatial heterogeneity of cooperators and non-cooperators. (A) Modeling simulation and (B) growth experiment of two ‘cooperator’ strains (labeled with red and green fluorescent proteins, resulting in yellow color) indicate a high degree of intermixing. (C) Modeling simulation and (D) growth experiment of the two ‘cooperator’ strains (red) and a ‘non-cooperator’ strain (green) indicate spatial segregation of non-cooperating bacteria. MALDI-TOF MS analysis of histidine and tryptophan concentrations in colonies illustrates the exclusion of these molecules from non-cooperator regions (reprinted with permission from Pande et al. 2016).
Similar articles
- Microbial Surface Colonization and Biofilm Development in Marine Environments.
Dang H, Lovell CR. Dang H, et al. Microbiol Mol Biol Rev. 2015 Dec 23;80(1):91-138. doi: 10.1128/MMBR.00037-15. Print 2016 Mar. Microbiol Mol Biol Rev. 2015. PMID: 26700108 Free PMC article. Review. - Social networks predict gut microbiome composition in wild baboons.
Tung J, Barreiro LB, Burns MB, Grenier JC, Lynch J, Grieneisen LE, Altmann J, Alberts SC, Blekhman R, Archie EA. Tung J, et al. Elife. 2015 Mar 16;4:e05224. doi: 10.7554/eLife.05224. Elife. 2015. PMID: 25774601 Free PMC article. - Inferring human microbial dynamics from temporal metagenomics data: Pitfalls and lessons.
Cao HT, Gibson TE, Bashan A, Liu YY. Cao HT, et al. Bioessays. 2017 Feb;39(2). doi: 10.1002/bies.201600188. Epub 2016 Dec 21. Bioessays. 2017. PMID: 28000336 Review. - High definition for systems biology of microbial communities: metagenomics gets genome-centric and strain-resolved.
Turaev D, Rattei T. Turaev D, et al. Curr Opin Biotechnol. 2016 Jun;39:174-181. doi: 10.1016/j.copbio.2016.04.011. Epub 2016 Apr 23. Curr Opin Biotechnol. 2016. PMID: 27115497 Review. - Functional metagenomics of methylotrophs.
Kalyuzhnaya MG, Beck DA, Chistoserdova L. Kalyuzhnaya MG, et al. Methods Enzymol. 2011;495:81-98. doi: 10.1016/B978-0-12-386905-0.00006-1. Methods Enzymol. 2011. PMID: 21419916
Cited by
- Metabolic Basis for Mutualism between Gut Bacteria and Its Impact on the Drosophila melanogaster Host.
Sommer AJ, Newell PD. Sommer AJ, et al. Appl Environ Microbiol. 2019 Jan 9;85(2):e01882-18. doi: 10.1128/AEM.01882-18. Print 2019 Jan 15. Appl Environ Microbiol. 2019. PMID: 30389767 Free PMC article. - Microbial active functional modules derived from network analysis and metabolic interactions decipher the complex microbiome assembly in mangrove sediments.
Du H, Pan J, Zou D, Huang Y, Liu Y, Li M. Du H, et al. Microbiome. 2022 Dec 13;10(1):224. doi: 10.1186/s40168-022-01421-w. Microbiome. 2022. PMID: 36510268 Free PMC article. - Interspecies Social Spreading: Interaction between Two Sessile Soil Bacteria Leads to Emergence of Surface Motility.
McCully LM, Bitzer AS, Seaton SC, Smith LM, Silby MW. McCully LM, et al. mSphere. 2019 Jan 30;4(1):e00696-18. doi: 10.1128/mSphere.00696-18. mSphere. 2019. PMID: 30700513 Free PMC article. - Nutrients Changed the Assembly Processes of Profuse and Rare Microbial Communities in Coals.
Zhang Y, Xue S, Chang X, Li Y, Yue X. Zhang Y, et al. Pol J Microbiol. 2022 Sep 24;71(3):359-370. doi: 10.33073/pjm-2022-032. eCollection 2022 Sep 1. Pol J Microbiol. 2022. PMID: 36185017 Free PMC article. - Cofactor Selectivity in Methylmalonyl Coenzyme A Mutase, a Model Cobamide-Dependent Enzyme.
Sokolovskaya OM, Mok KC, Park JD, Tran JLA, Quanstrom KA, Taga ME. Sokolovskaya OM, et al. mBio. 2019 Sep 24;10(5):e01303-19. doi: 10.1128/mBio.01303-19. mBio. 2019. PMID: 31551329 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources