The endocytic pathway in microglia during health, aging and Alzheimer's disease - PubMed (original) (raw)
Review
The endocytic pathway in microglia during health, aging and Alzheimer's disease
Santiago Solé-Domènech et al. Ageing Res Rev. 2016 Dec.
Abstract
Microglia, the main phagocytes of the central nervous system (CNS), are involved in the surveillance and maintenance of nervous tissue. During normal tissue homeostasis, microglia migrates within the CNS, phagocytose dead cells and tissue debris, and modulate synapse pruning and spine formation via controlled phagocytosis. In the event of an invasion by a foreign body, microglia are able to phagocytose the invading pathogen and process it proteolytically for antigen presentation. Internalized substrates are incorporated and sorted within the endocytic pathway and thereafter transported via complex vesicular routes. When targeted for degradation, substrates are delivered to acidic late endosomes and lysosomes. In these, the enzymatic degradation relies on pH and enzyme content. Endocytosis, sorting, transport, compartment acidification and degradation are regulated by complex signaling mechanisms, and these may be altered during aging and pathology. In this review, we discuss the endocytic pathway in microglia, with insight into the mechanisms controlling lysosomal biogenesis and pH regulation. We also discuss microglial lysosome function associated with Alzheimer's disease (AD) and the mechanisms of amyloid-beta (Aβ) internalization and degradation. Finally, we explore some therapies currently being investigated to treat AD and their effects on microglial response to Aβ, with insight in those involving enhancement of lysosomal function.
Keywords: Alzheimer’s disease; Amyloid-beta; Endocytosis; Lysosome; MITF; Microglia; TFEB.
Copyright © 2016 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of Interests The authors declared no potential conflict of interests regarding the publication of this manuscript.
Figures
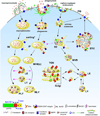
Figure 1. Proposed endocytic pathways in phagocytic cells
Endocytic mechanisms, namely macropinocytosis, phagocytosis and clathrin-mediated endocytosis, are depicted on the PM. Following receptor-mediated endocytosis, substrates bound to receptors undergo sorting in SEs and receptors are recycled back to the PM either directly from nascent endosomes or via the ERC, whereas nascent phagosomes and macropinosomes fuse with SEs and undergo a similar sorting process (indicated by an asterisk). SEs as well as intermediate phagosomes / macropinosomes containing sorted cargo (IPs and IMs, respectively) mature and adopt a multivesicular morphology (MVBs) where an additional sorting process takes place. Substrates in MVBs eventually reach LEs, which receive enzymes and membrane proteins from the TGN via vesicular trafficking, and the LY. Most degradation occurs in these acidic compartments. In contrast, IPs and IMs acquire enzymatic cargo by fusion with LEs and LYs, giving rise to phagolysosomes and macropinolysosomes. APP cellular trafficking: Surface APP is cleaved by α-secretase followed by extracellular release of the αAPP fragment. However, an important fraction of APP is incorporated into the endocytic pathway by clathrin-mediated endocytosis and processed mainly in SEs and the ERC by BACE1 and γ-secretase, leading to secretion of AICD to the cytosol and Aβ to the lumen of the compartments. Trafficking routes are indicated by black arrows. Fusion events are indicated by pink arrows. Abbreviations: PM, plasma membrane. SE, sorting endosome. ERC, endocytic recycling compartment. IP, intermediate phagosome. IM, intermediate macropinosome. MVB, multivesicular body. TGN, trans-Golgi network. LE, late endosome. LY, lysosome. PLY, phagolysosome. MLY, macropinolysosome. SRA, scavenger receptor class A. APP, amyloid precursor protein. αAPPs, soluble APP N-terminal fragment from α-secretase cleavage. βAPPs, soluble APP N-terminal fragment from β-secretase cleavage. AICD, APP intracellular domain. sAβ, soluble Aβ. fAβ, fibrillar Aβ. LDL, low density lipoprotein. M6PR, mannose-6-phosphate receptors.
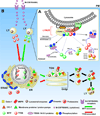
Figure 2. Proposed mechanisms of lysosomal biogenesis in microglia
(A) TFEB and MITF localization is dependent on their phosphorylation sites and states. In fully fed cells, TFEB (and probably MITF too) associates with active RAG GTPases in the LYNUS machinery at the surface of lysosomes and becomes phosphorylated by mTORC-1 at multiple sites including Ser211 and Ser142 (phosphorylation indicated by blue circles) followed by release to the cytosol and complex formation with 14-3-3 chaperones (indicated by black lines and arrowheads) (Martina and Puertollano, 2013; Settembre et al., 2013b). C-TAK1 and ERK1/2 have also been suggested to phosphorylate MITF and TFEB at Ser173 and Ser142, respectively, and promote their cytosolic stabilization by complex formation with 14-3-3 proteins (indicated by blue lines and arrowheads). Under certain conditions such as starvation or cellular stress, mTORC-1 detaches from LYNUS and no longer phosphorylates TFEB likely leading to destabilization of TFEB-14-3-3 complexes and promoting the nuclear translocation of the factor (indicated by grey lines and arrowheads). MITF may be regulated in a similar manner (Martina and Puertollano, 2013; Roczniak-Ferguson et al., 2012; Settembre et al., 2013b). In addition, following uptake by specific PM receptors, M-CSF and RANKL are also able to promote nuclear translocation of MITF by inhibiting C-TAK1 (which in turn destabilizes MITF-14-3-3 complexes) and/or promoting ERK 1/2-associated phosphorylation of the factor at Ser73 (Bronisz et al., 2006; Weilbaecher et al., 2001). RANKL alone has been shown to increase TFEB nuclear levels by preventing its degradation via PKC-β-associated phosphorylation at multiple serine residues (phosphorylation indicated by yellow circles), which seems to stabilize TFEB in the cytosol (Ferron et al., 2013). (B). Following nuclear mobilization, MITF and TFEB (depicted by green and red circles/arrows, respectively) promote the transcription of lysosomal genes (Meadows et al., 2007; Sardiello et al., 2009; Settembre et al., 2012). Newly synthesized lysosomal proteins pass through the ER and the Golgi and reach acidic late endosomes by vesicular trafficking (depicted by triangles, circles, rectangles and squares in the various organelles, and described in the legend). Abbreviations: M-CSF, macrophage-colony stimulating factor. RANKL, receptor activator of nuclear factor kappa-B ligand. LY, lysosome. MITF, microphthalmia-associated transcription factor. TFEB, transcription factor EB. C-TAK1, Cdc25C-associated kinase 1. MAPK, Mitogen-activated protein kinases. mTORC1, mammalian target of rapamycin complex 1. LYNUS, LY nutrient sensing machinery.
Similar articles
- Deacetylation of TFEB promotes fibrillar Aβ degradation by upregulating lysosomal biogenesis in microglia.
Bao J, Zheng L, Zhang Q, Li X, Zhang X, Li Z, Bai X, Zhang Z, Huo W, Zhao X, Shang S, Wang Q, Zhang C, Ji J. Bao J, et al. Protein Cell. 2016 Jun;7(6):417-33. doi: 10.1007/s13238-016-0269-2. Epub 2016 May 21. Protein Cell. 2016. PMID: 27209302 Free PMC article. - The Alzheimer's Disease Gene SORL1 Regulates Lysosome Function in Human Microglia.
Mishra S, Morshed N, Sidhu SB, Kinoshita C, Stevens B, Jayadev S, Young JE. Mishra S, et al. Glia. 2025 Jul;73(7):1329-1348. doi: 10.1002/glia.70009. Epub 2025 Apr 4. Glia. 2025. PMID: 40183375 Free PMC article. - Pseudoginsenoside-F11 alleviates oligomeric β-amyloid-induced endosome-lysosome defects in microglia.
Yao XC, Xue X, Zhang HT, Zhu MM, Yang XW, Wu CF, Yang JY. Yao XC, et al. Traffic. 2019 Jan;20(1):61-70. doi: 10.1111/tra.12620. Epub 2018 Dec 5. Traffic. 2019. PMID: 30375163 - Digestive exophagy as a novel mechanism of amyloid-β degradation by microglial lysosomes.
Meyer-Luehmann M. Meyer-Luehmann M. Trends Neurosci. 2025 Mar;48(3):174-175. doi: 10.1016/j.tins.2025.01.005. Epub 2025 Feb 13. Trends Neurosci. 2025. PMID: 39952833 Review. - Lysosomal acidification dysfunction in microglia: an emerging pathogenic mechanism of neuroinflammation and neurodegeneration.
Quick JD, Silva C, Wong JH, Lim KL, Reynolds R, Barron AM, Zeng J, Lo CH. Quick JD, et al. J Neuroinflammation. 2023 Aug 5;20(1):185. doi: 10.1186/s12974-023-02866-y. J Neuroinflammation. 2023. PMID: 37543564 Free PMC article. Review.
Cited by
- A multifaceted role of progranulin in regulating amyloid-beta dynamics and responses.
Du H, Wong MY, Zhang T, Santos MN, Hsu C, Zhang J, Yu H, Luo W, Hu F. Du H, et al. Life Sci Alliance. 2021 Jun 8;4(7):e202000874. doi: 10.26508/lsa.202000874. Print 2021 Jul. Life Sci Alliance. 2021. PMID: 34103390 Free PMC article. - Microglia specific deletion of miR-155 in Alzheimer's disease mouse models reduces amyloid-β pathology but causes hyperexcitability and seizures.
Aloi MS, Prater KE, Sánchez REA, Beck A, Pathan JL, Davidson S, Wilson A, Keene CD, de la Iglesia H, Jayadev S, Garden GA. Aloi MS, et al. J Neuroinflammation. 2023 Mar 7;20(1):60. doi: 10.1186/s12974-023-02745-6. J Neuroinflammation. 2023. PMID: 36879321 Free PMC article. - The Implication of Glial Metabotropic Glutamate Receptors in Alzheimer's Disease.
de Lima IBQ, Ribeiro FM. de Lima IBQ, et al. Curr Neuropharmacol. 2023;21(2):164-182. doi: 10.2174/1570159X20666211223140303. Curr Neuropharmacol. 2023. PMID: 34951388 Free PMC article. Review. - Role of Neuron and Glia in Alzheimer's Disease and Associated Vascular Dysfunction.
Bandyopadhyay S. Bandyopadhyay S. Front Aging Neurosci. 2021 Jun 15;13:653334. doi: 10.3389/fnagi.2021.653334. eCollection 2021. Front Aging Neurosci. 2021. PMID: 34211387 Free PMC article. - Inflammatory Processes in Alzheimer's Disease-Pathomechanism, Diagnosis and Treatment: A Review.
Twarowski B, Herbet M. Twarowski B, et al. Int J Mol Sci. 2023 Mar 30;24(7):6518. doi: 10.3390/ijms24076518. Int J Mol Sci. 2023. PMID: 37047492 Free PMC article. Review.
References
- Banati RB, Rothe G, Valet G, Kreutzberg GW. Detection of lysosomal cysteine proteinases in microglia: flow cytometric measurement and histochemical localization of cathepsin B and L. Glia. 1993;7:183–191. - PubMed
- Bard F, Cannon C, Barbour R, Burke RL, Games D, Grajeda H, Guido T, Hu K, Huang J, Johnson-Wood K, Khan K, Kholodenko D, Lee M, Lieberburg I, Motter R, Nguyen M, Soriano F, Vasquez N, Weiss K, Welch B, Seubert P, Schenk D, Yednock T. Peripherally administered antibodies against amyloid beta-peptide enter the central nervous system and reduce pathology in a mouse model of Alzheimer disease. Nat Med. 2000;6:916–919. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical