Understanding the evolution of holoparasitic plants: the complete plastid genome of the holoparasite Cytinus hypocistis (Cytinaceae) - PubMed (original) (raw)
Understanding the evolution of holoparasitic plants: the complete plastid genome of the holoparasite Cytinus hypocistis (Cytinaceae)
Cristina Roquet et al. Ann Bot. 2016.
Abstract
Background and Aims Plant plastid genomes are highly conserved in size, gene content and structure; however, parasitic plants are a noticeable exception to this evolutionary stability. Although the evolution of parasites could help to better understand plastome evolution in general, complete plastomes of parasites have been sequenced only for some lineages so far. Here we contribute to filling this gap by providing and analysing the complete plastome sequence of Cytinus hypocistis, the first parasite sequenced for Malvales and a species suspected to have an extremely small genome. Methods We sequenced and assembled de novo the plastid genome of Cytinus hypocistis using a shotgun approach on genomic DNA. Phylogenomic analyses based on coding regions were performed on Malvidae. For each coding region present in Cytinus, we tested for relaxation or intensification of selective pressures in the Cytinus lineage compared with autotrophic Malvales. Key Results Cytinus hypocistis has an extremely divergent genome that is among the smallest sequenced to date (19·4 kb), with only 23 genes and no inverted repeat regions. Phylogenomic analysis confirmed the position of Cytinus within Malvales. All coding regions of Cytinus plastome presented very high substitution rates compared with non-parasitic Malvales. Conclusions Some regions were inferred to be under relaxed negative selection in Cytinus, suggesting that further plastome reduction is occurring due to relaxed purifying selection associated with the loss of photosynthetic activity. On the other hand, increased selection intensity and strong positive selection were detected for rpl22 in the Cytinus lineage, which might indicate an evolutionary role in the host-parasite arms race, a point that needs further research.
Keywords: Chloroplast genome; Cytinaceae; Cytinus hypocistis; Malvales; mycoheterotroph; parasite; plastome evolution; selective pressure.
© The Author 2016. Published by Oxford University Press on behalf of the Annals of Botany Company. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
Fig. 1.
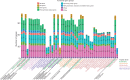
Number of unique plastid genes in each functional group of parasite and mycoheterotroph species and their closest autotrophic relative for which the whole plastome has been sequenced (Ptilidium: Forrest et al., 2011; Piper: Cai et al., 2006; Phalaenopsis: Chang et al., 2006; Dioscorea: Hansen et al., 2007; Carludovica: Lam et al., 2015; Vitis: Jansen et al., 2006; Ipomoea: McNeal et al., 2007; Lindenbergia: Wicke et al., 2013; Theobroma: Kane et al., 2012; all references concerning parasitic species are cited in the main text). The species are grouped by the family to which they belong (indicated in grey capital letters). The family of the autotrophic relative is indicated in lower-case letters when it belongs to a different family.
Fig. 2.
Chloroplast gene phylogeny of Malvidae: phylogram of the maximum likelihood tree determined by RAxML with 200 independent searches. Numbers associated with branches indicate bootstrap support values obtained with 1000 replicates; unnumbered branches had 100 % support. Scale indicates substitutions per site. Collapsed monophyletic clades correspond to five Oenothera, 20 Gossypium and 31 Eucalyptus species. The inserted image shows Cytinus hypocistis parasitizing a Cistus sp. (Stromboli, Aeolian Islands; © Cristina Roquet).
Fig. 3.
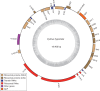
Chloroplast genome map of Cytinus hypocistis showing annotated genes. The grey circle indicates the GC content and the line marks the 50 % threshold. Ψ indicates pseudogenes and * indicates genes that might be pseudogenes.
Similar articles
- Mycoheterotrophic Epirixanthes (Polygalaceae) has a typical angiosperm mitogenome but unorthodox plastid genomes.
Petersen G, Darby H, Lam VKY, Pedersen HÆ, Merckx VSFT, Zervas A, Seberg O, Graham SW. Petersen G, et al. Ann Bot. 2019 Nov 15;124(5):791-807. doi: 10.1093/aob/mcz114. Ann Bot. 2019. PMID: 31346602 Free PMC article. - Do nonasterid holoparasitic flowering plants have plastid genomes?
Nickrent DL, Ouyang Y, Duff RJ, dePamphilis CW. Nickrent DL, et al. Plant Mol Biol. 1997 Jul;34(5):717-29. doi: 10.1023/a:1005860632601. Plant Mol Biol. 1997. PMID: 9278163 - Tannin profile, antioxidant properties, and antimicrobial activity of extracts from two Mediterranean species of parasitic plant Cytinus.
Maisetta G, Batoni G, Caboni P, Esin S, Rinaldi AC, Zucca P. Maisetta G, et al. BMC Complement Altern Med. 2019 Apr 5;19(1):82. doi: 10.1186/s12906-019-2487-7. BMC Complement Altern Med. 2019. PMID: 30952208 Free PMC article. - Cytinus under the Microscope: Disclosing the Secrets of a Parasitic Plant.
Sanjust E, Rinaldi AC. Sanjust E, et al. Plants (Basel). 2021 Jan 12;10(1):146. doi: 10.3390/plants10010146. Plants (Basel). 2021. PMID: 33445677 Free PMC article. Review. - The ins and outs of editing and splicing of plastid RNAs: lessons from parasitic plants.
Tillich M, Krause K. Tillich M, et al. N Biotechnol. 2010 Jul 31;27(3):256-66. doi: 10.1016/j.nbt.2010.02.020. Epub 2010 Mar 3. N Biotechnol. 2010. PMID: 20206308 Review.
Cited by
- Mycoheterotrophic Epirixanthes (Polygalaceae) has a typical angiosperm mitogenome but unorthodox plastid genomes.
Petersen G, Darby H, Lam VKY, Pedersen HÆ, Merckx VSFT, Zervas A, Seberg O, Graham SW. Petersen G, et al. Ann Bot. 2019 Nov 15;124(5):791-807. doi: 10.1093/aob/mcz114. Ann Bot. 2019. PMID: 31346602 Free PMC article. - Comparative Plastome Analysis of Root- and Stem-Feeding Parasites of Santalales Untangle the Footprints of Feeding Mode and Lifestyle Transitions.
Chen X, Fang D, Wu C, Liu B, Liu Y, Sahu SK, Song B, Yang S, Yang T, Wei J, Wang X, Zhang W, Xu Q, Wang H, Yuan L, Liao X, Chen L, Chen Z, Yuan F, Chang Y, Lu L, Yang H, Wang J, Xu X, Liu X, Wicke S, Liu H. Chen X, et al. Genome Biol Evol. 2020 Jan 1;12(1):3663-3676. doi: 10.1093/gbe/evz271. Genome Biol Evol. 2020. PMID: 31845987 Free PMC article. - Dense infraspecific sampling reveals rapid and independent trajectories of plastome degradation in a heterotrophic orchid complex.
Barrett CF, Wicke S, Sass C. Barrett CF, et al. New Phytol. 2018 May;218(3):1192-1204. doi: 10.1111/nph.15072. Epub 2018 Mar 4. New Phytol. 2018. PMID: 29502351 Free PMC article. - GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes.
Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. Jin JJ, et al. Genome Biol. 2020 Sep 10;21(1):241. doi: 10.1186/s13059-020-02154-5. Genome Biol. 2020. PMID: 32912315 Free PMC article. - Genetic, evolutionary and phylogenetic aspects of the plastome of annatto (Bixa orellana L.), the Amazonian commercial species of natural dyes.
Gomes Pacheco T, de Santana Lopes A, Monteiro Viana GD, Nascimento da Silva O, Morais da Silva G, do Nascimento Vieira L, Guerra MP, Nodari RO, Maltempi de Souza E, de Oliveira Pedrosa F, Otoni WC, Rogalski M. Gomes Pacheco T, et al. Planta. 2019 Feb;249(2):563-582. doi: 10.1007/s00425-018-3023-6. Epub 2018 Oct 11. Planta. 2019. PMID: 30310983
References
- Achaz G, Boyer F, Rocha EPC, Viari A, Coissac E. 2007. Repseek, a tool to retrieve approximate repeats from large DNA sequences. Bioinformatics 23: 119–121. - PubMed
- Barbrook AC, Howe CJ, Purton S. 2006. Why are plastid genomes retained in non-photosynthetic organisms? Trends in Plant Science 11: 101–108. - PubMed
- Barrett CF, Davis JI. 2012. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. American Journal of Botany 99: 1513–1523. - PubMed
- Barrett CF, Freudenstein JV, Li J, et al. 2014. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Molecular Biology and Evolution 31: 3095–3112. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous