Deciphering dynamics of clathrin-mediated endocytosis in a living organism - PubMed (original) (raw)
Deciphering dynamics of clathrin-mediated endocytosis in a living organism
Joshua P Ferguson et al. J Cell Biol. 2016.
Abstract
Current understanding of clathrin-mediated endocytosis (CME) dynamics is based on detection and tracking of fluorescently tagged clathrin coat components within cultured cells. Because of technical limitations inherent to detection and tracking of single fluorescent particles, CME dynamics is not characterized in vivo, so the effects of mechanical cues generated during development of multicellular organisms on formation and dissolution of clathrin-coated structures (CCSs) have not been directly observed. Here, we use growth rates of fluorescence signals obtained from short CCS intensity trace fragments to assess CME dynamics. This methodology does not rely on determining the complete lifespan of individual endocytic assemblies. Therefore, it allows for real-time monitoring of spatiotemporal changes in CME dynamics and is less prone to errors associated with particle detection and tracking. We validate the applicability of this approach to in vivo systems by demonstrating the reduction of CME dynamics during dorsal closure of Drosophila melanogaster embryos.
© 2016 Ferguson et al.
Figures
Figure 1.
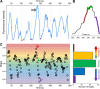
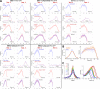
Determining CCS growth rate distributions. (A) Fluorescence intensity trace of a clathrin hotspot imaged at the ventral surface of a BSC-1 cell expressing AP2-GFP. Traces segments that are not used in growth rate calculation are shown in red, as they were considered the background signal. (B) Zoomed version of the clathrin-coated pit trace marked by the arrow in A. Shaded regions show 12-s-long fragments dwelling at formation (red), plateau (green), and dissolution (purple) phases of the pit. (C, left) For the intensity trace in A, slope values representing the growth rates are determined from 12-s-long fragments centered on each time point. Red, green, and purple arrowheads mark the slopes corresponding to the growth, plateau, and dissolution fragments highlighted in B, respectively. (C, right) Bar plot shows the distribution of the growth rates shown in the left panel. Positive and negative values correspond to formation and dissolution phases, respectively.
Figure 2.
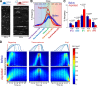
Using growth rate histograms as reporters of clathrin dynamics. (A) Kymographs are generated from the same BSC-1 cell before and during microaspiration, respectively. Elongated AP2 traces demonstrate longer CCS lifetimes under increased membrane tension. (B) Growth rate distributions are shown for seven different cells before and during aspiration. Change in CCS dynamics induced by micropipette aspiration can be observed in growth rate distributions. In the control experiments (cells before aspiration), CCSs spend more time in the formation phase (i.e., the distribution is inclined toward positive slopes). The asymmetry was abolished upon aspiration and plateau phases got relatively longer (_N_cells = 7 and _N_traces = 38136). (C) Growth rate distributions in B are assembled in five bins to better delineate different phases of clathrin-coated vesicle formation (FD, fast dissolution; FF, fast formation; P, plateau; SD, slow dissolution; SF, slow formation). The bars show mean + standard deviation to illustrate dispersion between cells. P-values were obtained using the two-tailed t test. (D) 2D histograms of normalized intensity traces aligned at different time points (beginning, trace maximum, and end) and superposed as represented by the cartoons. In each alignment, the aspirated cells show a significantly widened distribution, demonstrating a preference for slopes lower in magnitude. Bins corresponding to multiples of 12 s are more populated, as they contain trace data from both 3- and 4-s frame rate acquisitions.
Figure 3.
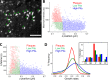
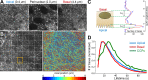
CCSs in proximity of substrate adhesion sites have reduced dynamics. (A) Arrowheads mark clathrin-coated plaques at the ventral surface of a BSC-1 cell stably expressing AP2-GFP. Bar, 5 µm. (B) Scatterplot shows maximum intensity versus the axial positions of CCSs detected at the ventral surface. The positions are relative to the lower bound of the confocal volume used for 3D tracking (Kural et al., 2012). CCS traces are divided into plaque (red) and pit (green and blue) populations based on their maximum intensities. Clathrin pits positioned closer to the substrate are in green (_N_cells = 6 and _N_traces = 11,482). (C) Scatterplot shows the lifetime versus axial positions of the CCSs shown in B. (D) Growth rate distributions of the pit and plaque populations. The inset shows the distributions as bar plots.
Figure 4.
Growth rate analysis does not depend on determining the complete traces of CCSs. (A, left) Thumbnails represent 12 temporal sections (24 s each) of a 288-s-long fluorescence movie of a BSC-1 cell stably expressing AP2-GFP. (right) CCS growth rate distribution of the entire movie. (B, left) Integrated movie segments (IMSs) are the quadrature sum of four 24-s-long segments in which density of CCSs is approximately four times larger than the original movie. Temporal separation is maximized between the segments to minimize the number of self-overlapping clathrin spots. (right) Growth rate distributions obtained from each IMS is plotted in comparison with the distribution obtained from the entire movie (black). (C) Growth rate distributions of the IMSs and the entire movie are displayed as bar plots. (D) We tested the reproducibility of growth rate distributions using the CCS traces produced by cmeAnalysis (Aguet et al., 2013) and TraCKer (homemade particle tracking program; see
online supplemental software
). Changes in slope distributions upon micropipette aspiration can be observed in both datasets. Numbers of the analyzed IMSs are 25 and 64 for before and during aspiration conditions, respectively (_N_cells = 7). Error bars show standard deviations. FD, fast dissolution; FF, fast formation; P, plateau; SD, slow dissolution; SF, slow formation.
Figure 5.
Real-time monitoring of CCS dynamics in cholesterol-depleted cells. (A) Fluorescence images show snapshots of three BSC-1 cells stably expressing AP2-GFP. 10 mM MβCD was applied at the sixth minute (t = 360 s) to initiate cholesterol depletion (
Video 2
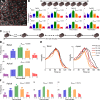
). Dashed lines demarcate the cell boundaries. Bar, 10 µm. (B) For the three cells shown in A, percentage frequency of the five growth phases are plotted as a function of time. The dashed lines denote the time point of MβCD addition. The fastest change in CCS dynamics is observed in cell 1. (C) Figures show standard deviation (of local clathrin growth rates for the snapshots shown in A. Each pixel in the image is given the value of standard deviation of the growth rates detected from CCSs found in a radius of 4.8 µm. This representation is used for illustrating temporal and spatial variations in CCS dynamics. Standard deviation is lower in regions where the growth rate distribution is narrow, a signature of impeded CCS dynamics. Cell 1 responds to cholesterol depletion the earliest, as observed at t = 816 s. (D) Growth rate histograms are shown for the three cells at the time points of the snapshots in A and C. Spatial and temporal heterogeneity of CCS dynamics can be detected using the standard deviation of growth rate histograms, which change at different rates for the three cells. (E) Plots show spatial variation in standard deviation of local CCS growth rates over the distances shown by the dashed arrows in C. Positions of the cell boundaries are marked by the dashed red lines.
Figure 6.
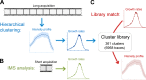
A novel analytical toolbox for CME dynamics. (A) CCS traces with similar intensity profiles can be grouped using a hierarchical clustering algorithm. This is applicable to acquisitions longer than the mean CCS lifetime. Growth rate distributions obtained from different clusters are used to develop a cluster library. (B) As validated by IMS analysis, growth rate distributions can be assembled using short fragments of CCS traces. (C) For a given growth rate distribution, an accurate estimation of the corresponding intensity profile is possible by determining analogous growth rate distributions in the cluster library.
Figure 7.
Determining CCS intensity profiles using growth rate distributions. (A) Trace clusters from test cells (excluded from the library) are matched with clusters from the CCS library by their growth rate histograms. Successful reproductions of the cluster traces occurred in multiple cell types and conditions. Two clusters and library matches are shown per condition. Intensity profiles of test clusters and corresponding growth rate distributions are shown in blue, and library matches are in red. Shaded regions in intensity profiles mark the standard deviation of the cluster. (B) In a U373 cell coexpressing fluorescently tagged AP2 and clathrin, the growth rate of AP2 is more peaked at low slope magnitudes (left). Corresponding library matches show continuing increase in clathrin signal after AP2 signal plateaued (right). (C) MβCD is added to BSC1 cells at t = 0. Growth rate distributions determined at different time points of the treatment are shown on the left. (right) Predicted intensity profiles for the given growth rate distributions. Gradual changes in growth rates result in increasing trace lifetime and peak intensity.
Figure 8.
3D tracking of CCSs in apical and basal surfaces of Drosophila amnioserosa. (A) Three sections of a confocal _z_-stack show apical, perinuclear, and basal regions of Drosophila amnioserosa cells expressing clathrin light chain fused with GFP (CLC-GFP). CCSs originating on organelles are observed mostly at the perinuclear regions as nondiffraction-limited bright fluorescent puncta (blobs). Bar, 10 µm. (B, left) Maximum projection image of a _z_-stack acquired at the amnioserosa tissue of a live Drosophila embryo. The image is divided into windows of 10 × 10 µm for determination of basal and apical surfaces (expanded in C). (right) Snapshot of CCS traces that are detected within the left panel, color-coded according to their axial positions. The projection of the entire 3D time-lapse acquisition and the corresponding CCS traces are shown in
Video 3
. (C) In each frame of the 3D time-lapse acquisition, axial positions of the apical and basal surfaces are determined separately for the 10 × 10 µm windows. Cartoon represents an amnioserosa cell oriented in a way that its apical surface faces the detection optics. The adjacent histogram shows the distribution of traces detected within the orange square shown in B with respect to their axial positions. The distribution is bimodal because of increased CCS density at the apical and basal surfaces and can be fit with a sum of two Gaussians. CCSs falling within one standard deviation of the respective means (± σ) are considered apical or basal. (D) A comparison of CCS lifetime distributions obtained from apical and basal amnioserosa with clathrin-coated pits (CCPs) detected in cultured BSC-1 cells stably expressing AP2-GFP (Amnioserosa: _N_embryos = 2, _N_cells = 75, and _N_traces = 124,013; CCPs: _N_cells = 3 and _N_traces = 12,002).
Figure 9.
CME dynamics in Drosophila amnioserosa. (A) Amnioserosa tissue of a late Drosophila embryo is imaged for 4 min using confocal _z_-stacks acquired every 3 s. Representative frame is an image section at the middle of a stack. Red lines represent the boundaries between amnioserosa cell centers, which are marked by numbers. Cell boundaries determined in each frame of a 3D time-lapse acquisition are shown in
Video 4
. Bar, 10 µm. (B) Histograms show evolution of the growth rates corresponding to different cells selected from the amnioserosa tissue in A. Frequencies of the five phases are plotted as a function of time for the apical and basal surfaces. (C) Thumbnails represent 30-s-long 3D time-lapse acquisitions separated by intermissions. (D) Transformation of CCS growth rates at the basal and apical surfaces of the amnioserosa during dorsal closure of a Drosophila embryo. Each bar in the histograms represents the frequency of growth rates obtained from CCS traces detected in individual 30-s-long acquisitions. A significant change in the growth rates is observed at the apical surface. (E) CCS intensity profiles predicted using the growth rates in (D). The change in the apical CCS dynamics is observed as gradually elongated lifetime (right). No major change is observed in the basal surface dynamics (left). (F) Histograms show the CCS growth rates at the apical amnioserosa of three embryos. Increasing frequency of the plateau phase is a hallmark of reduced CCS dynamics. FD, fast dissolution; FF, fast formation; P, plateau; SD, slow dissolution; SF, slow formation.
Similar articles
- Imaging clathrin dynamics in Drosophila melanogaster hemocytes reveals a role for actin in vesicle fission.
Kochubey O, Majumdar A, Klingauf J. Kochubey O, et al. Traffic. 2006 Dec;7(12):1614-27. doi: 10.1111/j.1600-0854.2006.00492.x. Epub 2006 Oct 2. Traffic. 2006. PMID: 17014698 - Membrane mechanics govern spatiotemporal heterogeneity of endocytic clathrin coat dynamics.
Willy NM, Ferguson JP, Huber SD, Heidotting SP, Aygün E, Wurm SA, Johnston-Halperin E, Poirier MG, Kural C. Willy NM, et al. Mol Biol Cell. 2017 Nov 15;28(24):3480-3488. doi: 10.1091/mbc.E17-05-0282. Epub 2017 Sep 13. Mol Biol Cell. 2017. PMID: 28904210 Free PMC article. - Visualizing clathrin-mediated endocytosis of G protein-coupled receptors at single-event resolution via TIRF microscopy.
Soohoo AL, Bowersox SL, Puthenveedu MA. Soohoo AL, et al. J Vis Exp. 2014 Oct 20;(92):e51805. doi: 10.3791/51805. J Vis Exp. 2014. PMID: 25350161 Free PMC article. - Mechanistic divergences of endocytic clathrin-coated vesicle formation in mammals, yeasts and plants.
Johnson A. Johnson A. J Cell Sci. 2024 Aug 15;137(16):jcs261847. doi: 10.1242/jcs.261847. Epub 2024 Aug 20. J Cell Sci. 2024. PMID: 39161994 Free PMC article. Review. - The long life of an endocytic patch that misses AP-2.
de León N, Valdivieso MH. de León N, et al. Curr Genet. 2016 Nov;62(4):765-770. doi: 10.1007/s00294-016-0605-3. Epub 2016 Apr 28. Curr Genet. 2016. PMID: 27126383 Review.
Cited by
- In Vivo Single-Molecule Tracking at the Drosophila Presynaptic Motor Nerve Terminal.
Bademosi AT, Lauwers E, Amor R, Verstreken P, van Swinderen B, Meunier FA. Bademosi AT, et al. J Vis Exp. 2018 Jan 14;(131):56952. doi: 10.3791/56952. J Vis Exp. 2018. PMID: 29364242 Free PMC article. - Nanoparticle Anisotropy Increases Targeting Interactions on Live-Cell Membranes.
Diloknawarit B, Lee K, Choo P, Odom TW. Diloknawarit B, et al. ACS Nano. 2024 May 14;18(19):12537-12546. doi: 10.1021/acsnano.4c02700. Epub 2024 Apr 29. ACS Nano. 2024. PMID: 38684051 - Load adaptation by endocytic actin networks.
Kaplan C, Kenny SJ, Chen X, Schöneberg J, Sitarska E, Diz-Muñoz A, Akamatsu M, Xu K, Drubin DG. Kaplan C, et al. Mol Biol Cell. 2022 May 15;33(6):ar50. doi: 10.1091/mbc.E21-11-0589. Epub 2022 Apr 7. Mol Biol Cell. 2022. PMID: 35389747 Free PMC article. - Cell Membrane Tension Gradients, Membrane Flows, and Cellular Processes.
Yan Q, Gomis Perez C, Karatekin E. Yan Q, et al. Physiology (Bethesda). 2024 Jul 1;39(4):0. doi: 10.1152/physiol.00007.2024. Epub 2024 Mar 19. Physiology (Bethesda). 2024. PMID: 38501962 Review. - Loss of PTEN promotes formation of signaling-capable clathrin-coated pits.
Rosselli-Murai LK, Yates JA, Yoshida S, Bourg J, Ho KKY, White M, Prisby J, Tan X, Altemus M, Bao L, Wu ZF, Veatch SL, Swanson JA, Merajver SD, Liu AP. Rosselli-Murai LK, et al. J Cell Sci. 2018 Apr 26;131(8):jcs208926. doi: 10.1242/jcs.208926. J Cell Sci. 2018. PMID: 29588397 Free PMC article.
References
- Apodaca G. 2002. Modulation of membrane traffic by mechanical stimuli. Am. J. Physiol. Renal Physiol. 282:F179–F190. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases