Antiparallel protocadherin homodimers use distinct affinity- and specificity-mediating regions in cadherin repeats 1-4 - PubMed (original) (raw)
Antiparallel protocadherin homodimers use distinct affinity- and specificity-mediating regions in cadherin repeats 1-4
John M Nicoludis et al. Elife. 2016.
Abstract
Protocadherins (Pcdhs) are cell adhesion and signaling proteins used by neurons to develop and maintain neuronal networks, relying on trans homophilic interactions between their extracellular cadherin (EC) repeat domains. We present the structure of the antiparallel EC1-4 homodimer of human PcdhγB3, a member of the γ subfamily of clustered Pcdhs. Structure and sequence comparisons of α, β, and γ clustered Pcdh isoforms illustrate that subfamilies encode specificity in distinct ways through diversification of loop region structure and composition in EC2 and EC3, which contains isoform-specific conservation of primarily polar residues. In contrast, the EC1/EC4 interface comprises hydrophobic interactions that provide non-selective dimerization affinity. Using sequence coevolution analysis, we found evidence for a similar antiparallel EC1-4 interaction in non-clustered Pcdh families. We thus deduce that the EC1-4 antiparallel homodimer is a general interaction strategy that evolved before the divergence of these distinct protocadherin families.
Keywords: biophysics; cell adhesion; computational biology; crystallography; human; protocadherins; sequence coevolution; structural biology; systems biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
Figure 1.. PcdhγB3 EC1-4 extended antiparallel dimer relies on unusual EC4 β-strand arrangement and is similar to other clustered Pcdh dimers.
(A) Structure of the PcdhγB3 EC1-4 antiparallel dimer, with each EC a different shade of blue and the Ca2+ ions in grey. (B) Superposition of PcdhγB3 EC2 and EC4 highlighting the differences in β-strands 1 and 2. (C) Comparison of the canonical cadherin (top) and EC4 (bottom) β-strand arrangement. (D) The structures of Pcdh dimers α4 EC1-4, α7 EC1-5, β6 EC1-4, and β8 EC1-4 (grey) were superimposed using the dimeric EC2-3 region onto γB3 EC1-4 (blue), illustrating variations in twist/corkscrew. (E) The EC4 domains of clustered Pcdh structures (colored as labeled) were superimposed, highlighting EC1 position differences that correlate with subfamily. Point of view (eye symbol) shown in (D). See Figure 1—figure supplements 1–4. DOI:
http://dx.doi.org/10.7554/eLife.18449.003
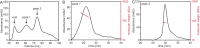
Figure 1—figure supplement 1.. PcdhγB3 refolding yields two species, one of which corresponds to monodisperse dimeric protein.
(A) SEC profile of refolded PcdhγB3 run on a Superdex 200 16/60 column, with the two collected peak fractions indicated. (B) SEC-MALS profile of Peak 1 run on a Superdex S200 10/300 column was broad and polydisperse. (C) SEC-MALS profile of Peak 2 run on a Superdex S200 10/300 column was monodisperse at a molecular weight of ~80 kDa, consistent with a dimer (monomeric molecular weight 47 kDa). DOI:
http://dx.doi.org/10.7554/eLife.18449.005
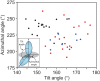
Figure 1—figure supplement 2.. Protocadherins and non-classical cadherins have a distribution of orientation between repeat pairs that is distinct from classical cadherins.
Distribution of tilt and azimuthal angles from adjacent EC repeat pairs of classical (black), Pcdh15 and Cdh23 (blue) and clustered Pcdh (red). (Inset) The orientation of adjacent EC repeats was defined by the tilt and azimuthal rotation of the EC domain principal axes. DOI:
http://dx.doi.org/10.7554/eLife.18449.006
Figure 1—figure supplement 3.. EC1 and EC3 use the same face for intersubunit contacts, as do EC2 and EC4.
(A) Sequence alignment of clustered Pcdhs for which dimer interface structures are available. EC1 and EC4 of PcdhγA1 are grey because their interaction interface is unknown. Residues highlighted orange have a BSA > 10 Å2 in those respective structures. We selected as conserved interface residues (boxed) those that have a BSA > 10 Å2 in 5 of 6 structures for EC2/EC3 or 4 of 5 structures for EC1/EC4. The interface regions are notably similar in EC1 and EC3, as well as in EC2 and EC4. (B) Superposition of the first half of PcdhγB3 EC1-4 with the second half yields an RMSD of 2.24 Å over 142 Cα atoms. (C) The two superimposed chains at the top (B) are rotated 90°, highlighting how similar surfaces of EC1 and EC3, and EC2 and EC4, form the extended antiparallel interface of clustered Pcdhs. In (B) and (C), the conserved interface positions identified in (A) are marked by Cα atom spheres. DOI:
http://dx.doi.org/10.7554/eLife.18449.007
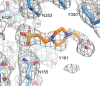
Figure 1—figure supplement 4.. HEPES molecule near the EC2/EC3 interface.
Final 2Fo-Fc electron density contoured at 1σ, with the final structural model shown as sticks. Nearby side chains from EC2 (lighter blue) and EC3 (darker blue) are labeled. DOI:
http://dx.doi.org/10.7554/eLife.18449.008
Figure 2.. Isoform-specific conservation and structural differences reveal subfamily differences in diversity generation.
(A) Subfamily-specific ICR values mapped onto the surfaces of Pcdhα7 (top, green), Pcdhβ8 (middle, magenta) and PcdhγB3 (bottom, blue). The black outline marks the dimer interface footprint. (B, C, D) Unique structural features of the α (left), β (center), and γ (right) structures (colored according to Figure 1). ICR values for highlighted residues shown below and normalized amino acid frequencies for these positions shown on the right. See Figure 2—figure supplement 1. DOI:
http://dx.doi.org/10.7554/eLife.18449.009
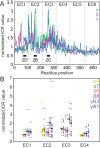
Figure 2—figure supplement 1.. Clustered Pcdh subfamilies have distinct patterns of isoform-specific conservation.
(A) Subfamily-specific normalized ICR values as a sliding average with a 5-residue window size (α = green, β = magenta, γ = cyan). Loop regions illustrated in Figure 2B–D are indicated at the bottom. (B) ICR values for interface residues (BSA > 10 Å2) for Pcdh α4 EC1-4 (yellow), α7 EC1-5 (green), β6 EC1-4 (salmon), β8 EC1-4 (magenta), γB3 EC1-4 (cyan), and γA1 EC1-3 (dark blue). In black are the average and standard error. Residues with high isoform-specific conservation localize to EC2 and EC3 surfaces. DOI:
http://dx.doi.org/10.7554/eLife.18449.010
Figure 3.. The EC1/EC4 interface is enriched in affinity-driving hydrophobic residues, while the EC2/EC3 interface contains high-ICR residues driving specificity.
(A) Amino acid frequencies in clustered Pcdhs of conserved interface residues (see Figure 1—figure supplement 2). (B) Plot of ICR value and ∆∆Gcalc of interface residues of Pcdh α4 EC1-4 (yellow), α7 EC1-5 (green), β6 EC1-4 (salmon), β8 EC1-4 (magenta), γB3 EC1-4 (blue). Two subsets of interface residues segregate from the main cluster: high-∆∆Gcalc and low-ICR residues (‘affinity’; black box) and low-∆∆Gcalc and high-ICR residues (‘specificity’; crimson box). Residue F86 from PcdhγB3 EC1-4 is labeled. (C) High-∆∆Gcalc and low-ICR residues (black) map primarily to EC1 and EC4, while low-∆∆Gcalc and high-ICR residues (crimson) primarily map to EC2 and EC3. N253 (*) is found in the ‘specificity’ region for γB3 and in the ‘affinity’ region for β6 and β8. (D) The EC1/EC4 interface features a hydrophobic cluster, with EC1 F86 near its center. (E) SEC-MALS profiles of WT PcdhγB3 EC1-4 (blue; molecular weight 82 kDa) and F86A (black; molecular weight 52 kDa) run on a Superdex S200 10/300 column, are consistent with dimeric and monomeric proteins, respectively. DOI:
http://dx.doi.org/10.7554/eLife.18449.011
Figure 4.. Evolutionary couplings in non-clustered Pcdhs predict an antiparallel interface engaging EC1-EC4.
(A) The top 38 covarying pairs are shown in black, and include a number of EC1-EC4 and EC2-EC3 covarying residue pairs. The intramolecular contact maps of PcdhγB3 EC1-4, Pcdhα4 EC1-4, Pcdhα7 EC1-4, Pcdhβ6 EC1-4, Pcdhβ8 EC1-4 and PcdhγA1 EC1-3 are in gray for reference. The observed interface contact residues are also mapped (α4, yellow; α7, green; β6, salmon; β8, magenta; γB3, blue; γA1, dark blue). (B) Covarying residue pairs across EC1-EC4 or EC2-EC3 are mapped onto the PcdhγB3 EC1-4 structure with a line between coupled residues. Alignments and evolutionary couplings in Figure 4—source data 1 and 2. (C) Amino acid frequencies at non-clustered Pcdh alignment positions corresponding to the conserved interface residue positions identified in clustered Pcdhs (Figure 1—figure supplement 2). See Figure 4—figure supplements 1 and 2. DOI:
http://dx.doi.org/10.7554/eLife.18449.012
Figure 4—figure supplement 1.. Evolutionary couplings in clustered Pcdhs are consistent with all available EC1-EC4 antiparallel homodimeric interfaces.
The top 83 covarying pairs are shown in black. The intramolecular contact maps of PcdhγB3 EC1-4, Pcdhα4 EC1-4, Pcdhα7 EC1-4, Pcdhβ6 EC1-4, Pcdhβ8 EC1-4 and PcdhγA1 EC1-3 are in gray for reference. The observed interface contact residues are also mapped (α4, yellow; α7, green; β6, salmon; β8, magenta; γB3, blue; γA1, dark blue). Alignments and evolutionary couplings in Figure 4—source data 3 and 4. DOI:
http://dx.doi.org/10.7554/eLife.18449.017
Figure 4—figure supplement 2.. Phylogenetic tree distinguishes clustered from non-clustered Pcdhs.
Based on this phylogeny, evolutionary couplings were obtained for the two groups labeled. The clustered and non-clustered Pcdh alignments had effective sequences numbers of 2660 and 405.5, respectively. DOI:
http://dx.doi.org/10.7554/eLife.18449.018
Similar articles
- Structure and Sequence Analyses of Clustered Protocadherins Reveal Antiparallel Interactions that Mediate Homophilic Specificity.
Nicoludis JM, Lau SY, Schärfe CP, Marks DS, Weihofen WA, Gaudet R. Nicoludis JM, et al. Structure. 2015 Nov 3;23(11):2087-98. doi: 10.1016/j.str.2015.09.005. Epub 2015 Oct 15. Structure. 2015. PMID: 26481813 Free PMC article. - Structural Basis of Diverse Homophilic Recognition by Clustered α- and β-Protocadherins.
Goodman KM, Rubinstein R, Thu CA, Bahna F, Mannepalli S, Ahlsén G, Rittenhouse C, Maniatis T, Honig B, Shapiro L. Goodman KM, et al. Neuron. 2016 May 18;90(4):709-23. doi: 10.1016/j.neuron.2016.04.004. Epub 2016 May 5. Neuron. 2016. PMID: 27161523 Free PMC article. - Combinatorial homophilic interaction between gamma-protocadherin multimers greatly expands the molecular diversity of cell adhesion.
Schreiner D, Weiner JA. Schreiner D, et al. Proc Natl Acad Sci U S A. 2010 Aug 17;107(33):14893-8. doi: 10.1073/pnas.1004526107. Epub 2010 Aug 2. Proc Natl Acad Sci U S A. 2010. PMID: 20679223 Free PMC article. - Non-clustered protocadherin.
Kim SY, Yasuda S, Tanaka H, Yamagata K, Kim H. Kim SY, et al. Cell Adh Migr. 2011 Mar-Apr;5(2):97-105. doi: 10.4161/cam.5.2.14374. Epub 2011 Mar 1. Cell Adh Migr. 2011. PMID: 21173574 Free PMC article. Review. - Structural origins of clustered protocadherin-mediated neuronal barcoding.
Rubinstein R, Goodman KM, Maniatis T, Shapiro L, Honig B. Rubinstein R, et al. Semin Cell Dev Biol. 2017 Sep;69:140-150. doi: 10.1016/j.semcdb.2017.07.023. Epub 2017 Jul 22. Semin Cell Dev Biol. 2017. PMID: 28743640 Free PMC article. Review.
Cited by
- Visualization of clustered protocadherin neuronal self-recognition complexes.
Brasch J, Goodman KM, Noble AJ, Rapp M, Mannepalli S, Bahna F, Dandey VP, Bepler T, Berger B, Maniatis T, Potter CS, Carragher B, Honig B, Shapiro L. Brasch J, et al. Nature. 2019 May;569(7755):280-283. doi: 10.1038/s41586-019-1089-3. Epub 2019 Apr 10. Nature. 2019. PMID: 30971825 Free PMC article. - Viscoelasticity, Like Forces, Plays a Role in Mechanotransduction.
Mierke CT. Mierke CT. Front Cell Dev Biol. 2022 Feb 9;10:789841. doi: 10.3389/fcell.2022.789841. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35223831 Free PMC article. Review. - Alpha protocadherins and Pyk2 kinase regulate cortical neuron migration and cytoskeletal dynamics via Rac1 GTPase and WAVE complex in mice.
Fan L, Lu Y, Shen X, Shao H, Suo L, Wu Q. Fan L, et al. Elife. 2018 Jun 18;7:e35242. doi: 10.7554/eLife.35242. Elife. 2018. PMID: 29911975 Free PMC article. - Chelicerata sDscam isoforms combine homophilic specificities to define unique cell recognition.
Zhou F, Cao G, Dai S, Li G, Li H, Ding Z, Hou S, Xu B, You W, Wiseglass G, Shi F, Yang X, Rubinstein R, Jin Y. Zhou F, et al. Proc Natl Acad Sci U S A. 2020 Oct 6;117(40):24813-24824. doi: 10.1073/pnas.1921983117. Epub 2020 Sep 22. Proc Natl Acad Sci U S A. 2020. PMID: 32963097 Free PMC article. - How clustered protocadherin binding specificity is tuned for neuronal self-/nonself-recognition.
Goodman KM, Katsamba PS, Rubinstein R, Ahlsén G, Bahna F, Mannepalli S, Dan H, Sampogna RV, Shapiro L, Honig B. Goodman KM, et al. Elife. 2022 Mar 7;11:e72416. doi: 10.7554/eLife.72416. Elife. 2022. PMID: 35253643 Free PMC article.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- P41 GM103403/GM/NIGMS NIH HHS/United States
- R01 GM106303/GM/NIGMS NIH HHS/United States
- S10 RR028832/RR/NCRR NIH HHS/United States
- S10 RR029205/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases