Classical pathology and mutational load of breast cancer - integration of two worlds - PubMed (original) (raw)
. 2015 Jul 20;1(4):225-38.
doi: 10.1002/cjp2.25. eCollection 2015 Oct.
Affiliations
- PMID: 27499907
- PMCID: PMC4939893
- DOI: 10.1002/cjp2.25
Classical pathology and mutational load of breast cancer - integration of two worlds
Jan Budczies et al. J Pathol Clin Res. 2015.
Abstract
Breast cancer is a complex molecular disease comprising several biological subtypes. However, daily routine diagnosis is still based on a small set of well-characterized clinico-pathological variables. Here, we try to link the two worlds of surgical pathology and multilayered molecular profiling by analyzing the relationships between clinico-pathological phenotypes and mutational loads of breast cancer. We evaluated the number of mutated genes with somatic non-silent mutations in different subgroups of breast cancer based on clinico-pathological, including immunohistochemical and tumour characteristics. The analysis was performed for a cohort of 687 primary breast cancer patients with mutational profiling, gene expression and clinico-pathological data available from The Cancer Genome Atlas (TCGA) project. The number of mutated genes was strongly positively associated with higher tumour grade (p = 1.4e-14) and with the different immunohistochemical and PAM50 molecular subtypes of breast cancer (p = 1.4e-10 and p = 4.3e-10, respectively). We observed significant associations (|R| > 0.4) between the abundance of mutated genes and expression levels of genes related to proliferation in the overall cohort and hormone receptor positive cohort, including the Recurrence Score gene signature (e.g., MYBL2 and BIRC5). Specific mutated genes (TP53, NCOR1, NF1, PTPRD and RB1) were highly significantly associated with high loads of mutated genes. Multivariate analysis for overall survival (OS) revealed a worse survival for patients with high numbers of mutated genes (hazard ratio = 4.6, 95% CI: 1.0 - 20.0, p = 0.044). Here, we report a strong association of the number of mutated genes with immunohistochemical and PAM50 subtypes and tumour grade in breast cancer. We provide evidence that specific levels of the mutational load underlie different morphological and biological phenotypes, which collectively constitute the current basis of pathological diagnosis. Our study is a step towards genomics-informed breast pathology and will provide a basis for future studies in this field bridging the gap between morphology, tumour biology and medical oncology.
Keywords: breast cancer; clinical parameters; genetics; mutations; pathology; prognosis; staging; tumour grade; tumour size.
Figures
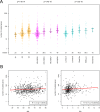
Figure 1
Association of the number of mutated genes (non‐silent somatic mutation) with clinico‐pathological characteristics of breast cancer. (A) In the beeswarm plot, each coloured dot represents a tumour. The bands indicate the first quartile, the median and the third quartile (n = median of mutated genes). The number of mutated genes was strongly associated with tumour grade, molecular subtyping by immunohistochemistry and SISH for ER, PR and HER2 as well as molecular subtyping by PAM50. (B) Scatterplots showing weak, but significant association of the number of mutations with patient age and tumour size in cm. The red curves represent the results of robust linear fitting. Not that the results of linear modeling appear as curves (not straight lines), because of the logarithmic scale of the _y_‐axis.
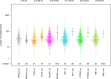
Figure 2
Association of the number of mutated genes (non‐silent somatic mutation) with mutation status of genes frequently mutated in breast cancer. In the beeswarm plot, each coloured dot represents a tumour. The bands indicate the median including the first and third quartile (n = median of mutated genes). The number of mutated genes was not associated with PIK3CA mutation status, but strongly and highly significantly associated with mutated TP53. Moreover, the number of mutated genes was strongly and significantly associated with mutated NCOR1, NF1, PTPRD and RB1.
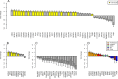
Figure 3
Gene expression FCs with 95% CIs between tumours with a high number of mutated genes (22 or more) and a low number of mutated genes (21 or less). Cell cycle genes [GO category 0000278: ‘mitotic cell cycle’; (40)] are colored in yellow. (A) Analysis of breast cancer: FCs of 44 genes whose expression strongly correlated with the number of mutated genes (|Spearman‐R|>0.4). (B) Analysis of HR+breast cancer: FCs of 11 genes whose expression strongly correlated with the number of mutated genes (|Spearman‐R|>0.4). (C) Analysis of HR−breast cancer: FCs of 23 genes whose expression strongly correlated with the number of mutated genes (|Spearman‐R|>0.4). (D) Analysis of HR+breast cancer: FCs of the 16 cancer genes included in the Recurrence Score assay.
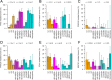
Figure 4
Association of the mutation status of specific genes with tumour grade and immunohistochemical as well as PAM50 subtypes (mutation rates including 95% confidence intervals). Genes with significant association after Bonferroni correction are shown (TP53, GATA3, CDKN1B, PIK3CA, CDH1, and MAP3K1). For example, the number of PIK3CA mutated tumours decreased from 54.3 via 36.7% to 18.7% with increasing tumour grade (G1, G2 and G3), while the number of TP53 mutated tumours increased from 5.4 via 15.4% to 52.4%. Mutation status of PIK3CA, TP53, GATA3, CDH1 and MAP3K1 correlated significantly with the molecular subtype. PIK3CA mutations were much less frequent in triple‐negative breast cancer (7.2%) compared with the other subtypes (all mutation rates >17.4%). TP53 mutation rate was much higher in triple‐negative (71.2%) and HR−/HER2+(69.6%) breast cancer compared with HR+/HER2−(16.3%) and HR+/HER2+(32.2%) breast cancer.
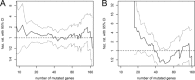
Figure 5
Association of overall survival with the total number of mutated genes (non‐silent somatic mutation) in breast cancer. Hazard ratios (haz. rat.) are shown for a dichotomized version of the number of mutated genes that was varied along the _x_‐axis. (A) In univariate analysis, no cut‐off point between 10 and 180 yielded a significant association with survival. (B) In multivariate analysis including correction for patient age, tumour size, nodal status, histopathological type, tumour grade, hormone receptor status and HER2 status, a high number of mutations were borderline‐significant associated with shorter survival for cut‐off points between 18.5 and 27.5. The most significant association was obtained for a cut‐off point of 21.5 with hazard ratio = 4.6 (1.0–10.0) and p = 0.044.
Similar articles
- PI3 kinase mutations and mutational load as poor prognostic markers in diffuse glioma patients.
Draaisma K, Wijnenga MM, Weenink B, Gao Y, Smid M, Robe P, van den Bent MJ, French PJ. Draaisma K, et al. Acta Neuropathol Commun. 2015 Dec 23;3:88. doi: 10.1186/s40478-015-0265-4. Acta Neuropathol Commun. 2015. PMID: 26699864 Free PMC article. - GATA3 mutations define a unique subtype of luminal-like breast cancer with improved survival.
Jiang YZ, Yu KD, Zuo WJ, Peng WT, Shao ZM. Jiang YZ, et al. Cancer. 2014 May 1;120(9):1329-37. doi: 10.1002/cncr.28566. Epub 2014 Jan 29. Cancer. 2014. PMID: 24477928 - Association of the germline TP53 R72P and MDM2 SNP309 variants with breast cancer survival in specific breast tumor subgroups.
van den Broek AJ, Broeks A, Horlings HM, Canisius SV, Braaf LM, Langerød A, Van't Veer LJ, Schmidt MK. van den Broek AJ, et al. Breast Cancer Res Treat. 2011 Nov;130(2):599-608. doi: 10.1007/s10549-011-1615-y. Epub 2011 Jun 11. Breast Cancer Res Treat. 2011. PMID: 21667122 - TP53 Mutational Analysis Enhances the Prognostic Accuracy of IHC4 and PAM50 Assays.
Lin CH, Chen IC, Huang CS, Hu FC, Kuo WH, Kuo KT, Wang CC, Wu PF, Chang DY, Wang MY, Chang CH, Chen WW, Lu YS, Cheng AL. Lin CH, et al. Sci Rep. 2015 Dec 16;5:17879. doi: 10.1038/srep17879. Sci Rep. 2015. PMID: 26671300 Free PMC article. - Oncogenic pathways in hereditary and sporadic breast cancer.
Kenemans P, Verstraeten RA, Verheijen RH. Kenemans P, et al. Maturitas. 2004 Sep 24;49(1):34-43. doi: 10.1016/j.maturitas.2004.06.005. Maturitas. 2004. PMID: 15351094 Review.
Cited by
- Mismatch repair testing in breast cancer: the path to tumor-specific immuno-oncology biomarkers.
Venetis K, Sajjadi E, Haricharan S, Fusco N. Venetis K, et al. Transl Cancer Res. 2020 Jul;9(7):4060-4064. doi: 10.21037/tcr-20-1852. Transl Cancer Res. 2020. PMID: 35117775 Free PMC article. No abstract available. - Immunotherapy in HER2-positive breast cancer: state of the art and future perspectives.
Krasniqi E, Barchiesi G, Pizzuti L, Mazzotta M, Venuti A, Maugeri-Saccà M, Sanguineti G, Massimiani G, Sergi D, Carpano S, Marchetti P, Tomao S, Gamucci T, De Maria R, Tomao F, Natoli C, Tinari N, Ciliberto G, Barba M, Vici P. Krasniqi E, et al. J Hematol Oncol. 2019 Oct 29;12(1):111. doi: 10.1186/s13045-019-0798-2. J Hematol Oncol. 2019. PMID: 31665051 Free PMC article. Review. - Knowledge mapping of tumor microenvironment for breast cancer: a bibliometric analysis from 2014 to 2023.
Jiang D, Wen P, Zhang S, Zhang N, Shao Q, Wang G, Wang L, Li S, Qin Y, Qu F, Zeng X. Jiang D, et al. Front Immunol. 2025 Mar 27;16:1550988. doi: 10.3389/fimmu.2025.1550988. eCollection 2025. Front Immunol. 2025. PMID: 40213549 Free PMC article. - The role of DNA damage repair (DDR) system in response to immune checkpoint inhibitor (ICI) therapy.
Shi C, Qin K, Lin A, Jiang A, Cheng Q, Liu Z, Zhang J, Luo P. Shi C, et al. J Exp Clin Cancer Res. 2022 Sep 7;41(1):268. doi: 10.1186/s13046-022-02469-0. J Exp Clin Cancer Res. 2022. PMID: 36071479 Free PMC article. Review. - Heterologous prime-boost cellular vaccination induces potent antitumor immunity against triple negative breast cancer.
Niavarani SR, St-Cyr G, Daniel L, Lawson C, Giguère H, Alkayyal AA, Tai LH. Niavarani SR, et al. Front Immunol. 2023 Feb 13;14:1098344. doi: 10.3389/fimmu.2023.1098344. eCollection 2023. Front Immunol. 2023. PMID: 36860852 Free PMC article.
References
- DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin 2014; 64: 52–62 - PubMed
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al Molecular portraits of human breast tumours. Nature 2000; 406: 747–752. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous