Genome-wide quantification of rare somatic mutations in normal human tissues using massively parallel sequencing - PubMed (original) (raw)
Genome-wide quantification of rare somatic mutations in normal human tissues using massively parallel sequencing
Margaret L Hoang et al. Proc Natl Acad Sci U S A. 2016.
Abstract
We present the bottleneck sequencing system (BotSeqS), a next-generation sequencing method that simultaneously quantifies rare somatic point mutations across the mitochondrial and nuclear genomes. BotSeqS combines molecular barcoding with a simple dilution step immediately before library amplification. We use BotSeqS to show age- and tissue-dependent accumulations of rare mutations and demonstrate that somatic mutational burden in normal human tissues can vary by several orders of magnitude, depending on biologic and environmental factors. We further show major differences between the mutational patterns of the mitochondrial and nuclear genomes in normal tissues. Lastly, the mutation spectra of normal tissues were different from each other, but similar to those of the cancers that arose in them. This technology can provide insights into the number and nature of genetic alterations in normal tissues and can be used to address a variety of fundamental questions about the genomes of diseased tissues.
Keywords: aging; genomics; next-generation sequencing; somatic mutation.
Conflict of interest statement
B.V. has no conflict of interest with respect to the new technology described in this manuscript, as defined by Johns Hopkins University's policy on conflict of interest. B.V. is a founder of PapGene and Personal Genome Diagnostics and a member of the Scientific Advisory Boards of Morphotek and Syxmex-Inostics. These companies and others have licensed patent applications on genetic technologies from Johns Hopkins, some of which result in royalty payments to B.V. The terms of these arrangements are being managed by Johns Hopkins University in accordance with its conflict of interest policies.
Figures
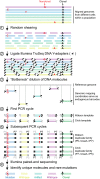
Fig. 1.
Bottleneck sequencing methodology. Each color at the top of the figure represents double-stranded DNA from a genome of one cell within a population. Random, nonclonal point mutations (red) are private to individual cells. In contrast, clonal reference changes (A in black) are present in all genomes within the cell population. (step 1) Random shearing generates variably sized DNA molecules. (step 2) Noncomplementary single-stranded regions of the Illumina Y-adapters (P5 in gray and P7 in black) are represented as forked structures ligated to both ends of each DNA molecule. (step 3) Dilution decreases the number of DNA molecules (five are shown) from the original population in a random manner. Ends of the DNA molecules align uniquely to the reference genome. Mapping coordinates are used as unique molecule “barcodes” during data processing. (step 4) PCR primer (black arrowhead) anneals and primer extends (hashed lines) the Watson and Crick template of the original DNA molecule independently. The red asterisk represents an error generated during PCR of the library. (step 5) Watson and Crick templates generate two families of PCR duplicates. Orientation of P5 (gray) and P7 (black) containing adapters to the DNA molecule (insert) distinguishes the two families. P5 and P7 sequences dictate which end will be sequenced in read 1 vs. read 2, respectively, on the Illumina flow cell. Red asterisks represent the PCR error propagated in the Watson but not the Crick family members. In contrast to artifacts, real mutations (C:G mutation in red) will be present in both the Watson and Crick family members. (step 6) The BotSeqS pipeline identifies and quantifies the number of unique DNA molecules and point mutations (C:G in red) in the sequencing data by eliminating artifacts and clonal changes (A:T in black).
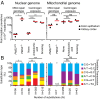
Fig. 2.
Nuclear point mutations increase in normal tissues from individuals with defects in DNA repair or with exposure to environmental carcinogens compared with controls. (A) Comparison of point mutation prevalences in nuclear (Left) and mitochondrial (Right) genome in age-matched normal colon epithelium (filled circle) with different DNA mismatch repair genotypes (PMS2 +/+ or PMS2 −/−) or in age-matched normal kidney cortex (filled square) without (none) or with (aristolochic acid or smoking) carcinogen exposure. Red lines represent average. *P < 0.05, _t_ test; **_P_ < 0.001 and ***_P_ < 0.0001, one-way ANOVA with Bonferroni multiple comparison posttest; ns, not significant, indicates _P_ > 0.05. (B) Stacked columns representing the substitution frequencies (y axis) of each substitution out of the six possible types (see legend). Cohort labels are indicated in A directly above each column. Number of substitutions (N) generating each mutational spectrum is indicated on the x axis. n.d., not determined due to an insufficient number of mutations (N = 7) for mutational spectrum analysis. *P = 0.04, Fisher’s exact test; **P = 2.6 × 10−8 and ***P = 1.5 × 10−16, Fisher’s exact test with Bonferroni multiple comparison correction; ns, not significant, indicates P > 0.05. All statistical tests in this figure were two-tailed.
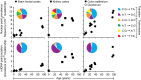
Fig. 3.
Normal human tissues accumulate point mutations over a lifetime with genome-specific and tissue-specific mutational patterns. Point mutation prevalences in nuclear (Top) and mitochondrial (Bottom) genome measured in four normal tissue types (brain frontal cortex of 9 individuals, kidney cortex of 5 individuals, colon epithelium of 11 individuals, and duodenum of 1 individual). Twenty-six total individuals were assessed, with each individual contributing to one normal tissue type. Pie chart Insets show the prevalences of each substitution out of the six possible substitution types (see pie chart legend, right side). Each pie chart was compiled from the individuals represented in their respective scatter plots, with the exception that duodenum was omitted. The number of substitutions generating the pie charts for the nuclear genome was n = 31 for brain, n = 73 for kidney, and n = 94 for colon, and for the mitochondrial genome was n = 181 for brain, n = 299 for kidney, and n = 116 for colon.
Similar articles
- Genome-wide mitochondrial DNA sequence variations and lower expression of OXPHOS genes predict mitochondrial dysfunction in oral cancer tissue.
Chattopadhyay E, De Sarkar N, Singh R, Ray A, Roy R, Paul RR, Pal M, Ghose S, Ghosh S, Kabiraj D, Banerjee R, Roy B. Chattopadhyay E, et al. Tumour Biol. 2016 Sep;37(9):11861-11871. doi: 10.1007/s13277-016-5026-x. Epub 2016 Apr 7. Tumour Biol. 2016. PMID: 27055661 - Comprehensive Mitochondrial Genome Analysis by Massively Parallel Sequencing.
Palculict ME, Zhang VW, Wong LJ, Wang J. Palculict ME, et al. Methods Mol Biol. 2016;1351:3-17. doi: 10.1007/978-1-4939-3040-1_1. Methods Mol Biol. 2016. PMID: 26530670 - Migration of mitochondrial DNA in the nuclear genome of colorectal adenocarcinoma.
Srinivasainagendra V, Sandel MW, Singh B, Sundaresan A, Mooga VP, Bajpai P, Tiwari HK, Singh KK. Srinivasainagendra V, et al. Genome Med. 2017 Mar 29;9(1):31. doi: 10.1186/s13073-017-0420-6. Genome Med. 2017. PMID: 28356157 Free PMC article. - Next-Generation Sequencing.
Le Gallo M, Lozy F, Bell DW. Le Gallo M, et al. Adv Exp Med Biol. 2017;943:119-148. doi: 10.1007/978-3-319-43139-0_5. Adv Exp Med Biol. 2017. PMID: 27910067 Review. - Emerging patterns of somatic mutations in cancer.
Watson IR, Takahashi K, Futreal PA, Chin L. Watson IR, et al. Nat Rev Genet. 2013 Oct;14(10):703-18. doi: 10.1038/nrg3539. Epub 2013 Sep 11. Nat Rev Genet. 2013. PMID: 24022702 Free PMC article. Review.
Cited by
- A single-stranded based library preparation method for virome characterization.
Zhai X, Gobbi A, Kot W, Krych L, Nielsen DS, Deng L. Zhai X, et al. Microbiome. 2024 Oct 24;12(1):219. doi: 10.1186/s40168-024-01935-5. Microbiome. 2024. PMID: 39449043 Free PMC article. - Clonal hematopoiesis associated with epigenetic aging and clinical outcomes.
Nachun D, Lu AT, Bick AG, Natarajan P, Weinstock J, Szeto MD, Kathiresan S, Abecasis G, Taylor KD, Guo X, Tracy R, Durda P, Liu Y, Johnson C, Rich SS, Van Den Berg D, Laurie C, Blackwell T, Papanicolaou GJ, Correa A, Raffield LM, Johnson AD, Murabito J, Manson JE, Desai P, Kooperberg C, Assimes TL, Levy D, Rotter JI, Reiner AP, Whitsel EA, Wilson JG, Horvath S, Jaiswal S; NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium. Nachun D, et al. Aging Cell. 2021 Jun;20(6):e13366. doi: 10.1111/acel.13366. Epub 2021 May 29. Aging Cell. 2021. PMID: 34050697 Free PMC article. - Clonal hematopoiesis and nonhematologic disorders.
Jaiswal S. Jaiswal S. Blood. 2020 Oct 1;136(14):1606-1614. doi: 10.1182/blood.2019000989. Blood. 2020. PMID: 32736379 Free PMC article. Review. - Molecular origins of APOBEC-associated mutations in cancer.
Petljak M, Maciejowski J. Petljak M, et al. DNA Repair (Amst). 2020 Oct;94:102905. doi: 10.1016/j.dnarep.2020.102905. Epub 2020 Jul 6. DNA Repair (Amst). 2020. PMID: 32818816 Free PMC article. Review. - Rare Event Detection Using Error-corrected DNA and RNA Sequencing.
Wong WH, Tong RS, Young AL, Druley TE. Wong WH, et al. J Vis Exp. 2018 Aug 3;(138):57509. doi: 10.3791/57509. J Vis Exp. 2018. PMID: 30124656 Free PMC article.
References
- Albertini RJ, Nicklas JA, O’Neill JP, Robison SH. In vivo somatic mutations in humans: Measurement and analysis. Annu Rev Genet. 1990;24:305–326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 CA062924/CA/NCI NIH HHS/United States
- R01 CA057345/CA/NCI NIH HHS/United States
- R37 CA043460/CA/NCI NIH HHS/United States
- R37 CA057345/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources