Dissecting the precise role of H3K9 methylation in crosstalk with DNA maintenance methylation in mammals - PubMed (original) (raw)
doi: 10.1038/ncomms12464.
Jiqin Zhang 1, Ruoyu Chen 1, Lina Wang 1, Bo Li 1, Hao Cheng 2, Xiaoya Duan 1, Haijun Zhu 1, Wei Wei 1, Jiwen Li 1, Qihan Wu 1, Jing-Dong J Han 2, Wenqiang Yu 3, Shaorong Gao 4, Guohong Li 5, Jiemin Wong 1 6
Affiliations
- PMID: 27554592
- PMCID: PMC5426519
- DOI: 10.1038/ncomms12464
Dissecting the precise role of H3K9 methylation in crosstalk with DNA maintenance methylation in mammals
Qian Zhao et al. Nat Commun. 2016.
Abstract
In mammals it is unclear if UHRF1-mediated DNA maintenance methylation by DNMT1 is strictly dependent on histone H3K9 methylation. Here we have generated an Uhrf1 knockin (KI) mouse model that specifically abolishes the H3K9me2/3-binding activity of Uhrf1. The homozygous Uhrf1 KI mice are viable and fertile, and exhibit ∼10% reduction of DNA methylation in various tissues. The reduced DNA methylation occurs globally in the genome and does not restrict only to the H3K9me2/3 enriched repetitive sequences. In vitro UHRF1 binds with higher affinity to reconstituted nucleosome with hemi-methylated CpGs than that with H3K9me2/3, although it binds cooperatively to nucleosome with both modifications. We also show that the nucleosome positioning affects the binding of methylated DNA by UHRF1. Thus, while our study supports a role for H3K9 methylation in promoting DNA methylation, it demonstrates for the first time that DNA maintenance methylation in mammals is largely independent of H3K9 methylation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
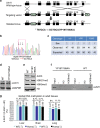
Figure 1. Generation and characterization of Uhrf1 YP187/188AA KI mice.
(a) Diagram illustrating the scheme for generation of the Uhrf1 YP187/188AA KI mice. The targeting vector contains Neo gene flanked by two FRT sites (triangles) and thymidine kinase (TK) gene for double selection. The asterisk represents the mutations of YP187/188AA (TATCCA→GCTGCA). (b) A representative sequencing data for homozygous YP187/188AA KI mice. (c) Summary of the genotyping results from the breeding of Uhrf1 KI heterozygous mice. (d) Western blot result showing the levels of Uhrf1 proteins in liver tissues from the WT and KI mice. (e) Western blot analysis of core histones prepared from liver tissues of the WT and KI mice using antibodies as indicated. (f) In vitro pulldown assay confirmed that the Uhrf1 proteins from the liver tissues of KI mice were impaired in binding the histone N-terminal tail peptide with H3K9me3. (g) The global levels of DNA methylation in various mouse tissues derived from the WT and homozygous KI mice determined by HPLC analysis. Tissues were derived from three pairs of WT and KI littermates. The resulting genomic DNAs were pooled together for HPLC analysis. The level of methylation was shown as the percentage of 5dmC to 5dmC+5dC.
Figure 2. Genome-wide analysis of DNA methylation by RRBS demonstrates that the YP187/188AA mutation results in a global reduction of DNA methylation.
(a) RRBS reads were mapped to mouse genome mm9 by bismark v0.12.2. Only unique mapped reads were kept for further analysis. CpGs and non CpGs in both strands were reported. Differentially methylated CpGs were obtained by using R package methylkit and only the CpGs with at least 5 reads coverage were used for further analysis. Differentially methylated CpGs and non CpGs were defined by FDR<0.05 and the absolute methylation difference above 25%. (b) 2,654,195 CpGs with at least 5 reads coverage in both WT and KI samples were plotted based on the percentages of DNA methylation. Mean CpG methylation levels in WT and KI are 45.6 and 42.6, respectively. Median CpG methylation levels in WT and KI are 46.7 and 38, respectively. Pair-wise _t_-test showed that the WT CpG methylation level was significantly higher than KI (P value<0.001). (c) The distribution of CpGs with different percentage of methylation levels in WT and Uhrf1 KI. 2,654,195 CpGs covered by both samples were analysed here. X axis is the percentage of DNA methylation, Y axis is CpG counts in million. (d) The methylation difference between KI and WT for CpGs in each quantile was calculated and plotted in boxplot. Highly methylated CpG sites (above 70%) decreased more, but decrease of methylation was seen for each quantile of CpG methylation. But only 2,512 CpGs were found with high methylation levels in WT (above 70%) and low methylation levels in KI (below 20%). (e) Average CpG methylation levels in WT and KI from 2 kb upstream and 2 kb downstream of gene transcriptional start sites. (f) Methylation differences in various repeat sequences were shown in boxplots. Each box represents one repeat class methylation difference. 304,709 single repeat elements were used with at least one CpG covered. Only 137 single repeat elements were found with high methylation levels in WT (above 70%) and low methylation levels in KI (below 20%).
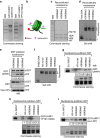
Figure 3. UHRF1 preferentially binds in vitro reconstituted nucleosome containing H3K9me3.
(a) Commassie blue staining gel showing recombinant UHRF1 fusion proteins. The recombinant GST-UHRF1 (aa 95–610) and its YP191/192AA mutant were expressed and purified from E.coli. The YP191/192AA mutation in human UHRF1 was equivalent to the YP187/188AA mutation in mouse Uhrf1. (b) Diagram illustrating the in vitro reconstituted nucleosome with histone octamers containing with (or without) H3K9me2/3-containing H3. The 200 bp 601 sequence with a biotin at one end was used for in vitro nucleosome assembly via salt dialysis. (c) Verification of the histone composition of in vitro reconstituted nucleosomes with different H3K9 methylation status by Commassie blue staining gel. (d) Verification of in vitro reconstituted nucleosomes with different H3K9 methylation status by gel mobility shift assay. Asterisk marks free DNA. (e) The reconstituted nucleosomes were tested for binding to immobilized GST-UHRF1 by pulldown assay. Note only the nucleosomes with H3K9me2 or H3K9me3 bound to GST-UHRF1 but not the control GST. (f) Gel mobility assay examining the nucleosomes derived from in vitro assembly with histone octamers containing H3 without N-terminal tail or different levels of H3K9 methylation. (g) The nucleosomes assembled in (f) were immobilized to streptavidin agarose beads and assayed for binding of GST-UHRF1. The Commassie staining gel at lower panel showed the compositions of core histones in corresponding in vitro assembled nucleosomes. (h) and (i) The pulldown was performed as in (g) except the GST-UHRF1 YP191/192AA mutant proteins and GST-UHRF1 (aa 1–407) without SRA domain were used, respectively.
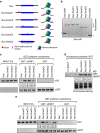
Figure 4. UHRF1 binds nucleosomes with hemi-methylated DNA and the binding is influenced by the nucleosome positioning of hemi-methylated CpGs.
(a) Diagram illustrating six different types of in vitro reconstituted nucleosomes with hemi-methylated CpG at distinct positions. The DNA fragments for Nuc, Nuc-5meC1, Nuc-5meC2 and Nuc-5meC3 were 200 bp with the 146 bp 601 sequence at the middle, whereas the DNA fragments for Nuc-5meC4 and Nuc-5meC5 were 163 bp. The relative positions and number of hemi-methylated CpGs were shown as.. (b) Gel mobility shift assay verified the in vitro reconstituted nucleosomes. Asterisk marks free DNA. (c) Immobilized GST-UHRF1 was used to pulldown in vitro reconstituted nucleosomes with hemi-methylated CpGs outside or within the edge of nucleosomes. (d) In vitro reconstituted nucleosomes as in (c) were immobilized to streptavidin agarose beads and used to pulldown GST-UHRF1 proteins. (e) Immobilized GST-UHRF1 was used to pulldown in vitro reconstituted nucleosomes with hemi-methylated CpGs outside, within the edge or near the dyad of nucleosomes.
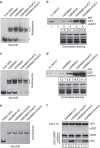
Figure 5. H3K9me3 and hemi-methylated CpGs cooperatively enhance the binding of UHRF1 to nucleosomes.
(a) Gel mobility shift assay showing the nucleosomes reconstituted with histone octamers containing with or without H3K9me3 and 200 bp DNA fragment with or without hemi-methylated CpGs outside of the nucleosome. (b) In vitro reconstituted nucleosomes as in (a) were immobilized to streptavidin agarose beads and used to pulldown GST-UHRF1 proteins. (c) Gel mobility shift assay showing the nucleosomes reconstituted with histone octamers containing with or without H3K9me3 and 200 bp DNA fragment with or without hemi-methylated CpGs near the edge of nucleosome. (d) In vitro reconstituted nucleosomes as in (c) were immobilized to streptavidin agarose beads and used to pulldown GST-UHRF1 proteins. (e) Gel mobility shift assay showing the nucleosomes reconstituted with 163 bp DNA fragments with hemi-methylated CpGs at the edge or near the dyad of nucleosomes and histone octamers with or without H3K9me3. (f) GST-UHRF1 was immobilized and used for pulldown of In vitro reconstituted nucleosomes as in (e). Note that the binding of UHRF1 to the hemi-methylated CpGs near the dyad of nucleosome was substantially inhibited in comparison with the sites near the edge of nucleosome and that presence of H3K9me3 in nucleosome partially restored the binding of UHRF1.
Similar articles
- UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9.
Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J, Koseki H, Wong J. Liu X, et al. Nat Commun. 2013;4:1563. doi: 10.1038/ncomms2562. Nat Commun. 2013. PMID: 23463006 - S phase-dependent interaction with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA methylation maintenance.
Zhang J, Gao Q, Li P, Liu X, Jia Y, Wu W, Li J, Dong S, Koseki H, Wong J. Zhang J, et al. Cell Res. 2011 Dec;21(12):1723-39. doi: 10.1038/cr.2011.176. Epub 2011 Nov 8. Cell Res. 2011. PMID: 22064703 Free PMC article. - Non-germ Line Restoration of Genomic Imprinting for a Small Subset of Imprinted Genes in Ubiquitin-like PHD and RING Finger Domain-Containing 1 (Uhrf1) Null Mouse Embryonic Stem Cells.
Qi S, Wang Z, Li P, Wu Q, Shi T, Li J, Wong J. Qi S, et al. J Biol Chem. 2015 May 29;290(22):14181-91. doi: 10.1074/jbc.M114.626697. Epub 2015 Apr 21. J Biol Chem. 2015. PMID: 25900245 Free PMC article. - Regulation of maintenance DNA methylation via histone ubiquitylation.
Nishiyama A, Yamaguchi L, Nakanishi M. Nishiyama A, et al. J Biochem. 2016 Jan;159(1):9-15. doi: 10.1093/jb/mvv113. Epub 2015 Nov 20. J Biochem. 2016. PMID: 26590302 Free PMC article. Review. - Mechanisms of Inheritance of Chromatin States: From Yeast to Human.
Madhani HD. Madhani HD. Annu Rev Biophys. 2025 May;54(1):59-79. doi: 10.1146/annurev-biophys-070524-091904. Epub 2024 Dec 23. Annu Rev Biophys. 2025. PMID: 39715046 Review.
Cited by
- The Arabidopsis APOLO and human UPAT sequence-unrelated long noncoding RNAs can modulate DNA and histone methylation machineries in plants.
Fonouni-Farde C, Christ A, Blein T, Legascue MF, Ferrero L, Moison M, Lucero L, Ramírez-Prado JS, Latrasse D, Gonzalez D, Benhamed M, Quadrana L, Crespi M, Ariel F. Fonouni-Farde C, et al. Genome Biol. 2022 Aug 29;23(1):181. doi: 10.1186/s13059-022-02750-7. Genome Biol. 2022. PMID: 36038910 Free PMC article. - The multi-functionality of UHRF1: epigenome maintenance and preservation of genome integrity.
Mancini M, Magnani E, Macchi F, Bonapace IM. Mancini M, et al. Nucleic Acids Res. 2021 Jun 21;49(11):6053-6068. doi: 10.1093/nar/gkab293. Nucleic Acids Res. 2021. PMID: 33939809 Free PMC article. Review. - Histone H3K9 Trimethylation Downregulates the Expression of Brain-Derived Neurotrophic Factor in the Dorsal Hippocampus and Impairs Memory Formation During Anaesthesia and Surgery.
Wu T, Sun XY, Yang X, Liu L, Tong K, Gao Y, Hao JR, Cao J, Gao C. Wu T, et al. Front Mol Neurosci. 2019 Oct 25;12:246. doi: 10.3389/fnmol.2019.00246. eCollection 2019. Front Mol Neurosci. 2019. PMID: 31708739 Free PMC article. - Combinatorial quantification of 5mC and 5hmC at individual CpG dyads and the transcriptome in single cells reveals modulators of DNA methylation maintenance fidelity.
Chialastri A, Sarkar S, Schauer EE, Lamba S, Dey SS. Chialastri A, et al. Nat Struct Mol Biol. 2024 Aug;31(8):1296-1308. doi: 10.1038/s41594-024-01291-w. Epub 2024 Apr 26. Nat Struct Mol Biol. 2024. PMID: 38671229 - Inhibition of 11β-HSD1 Ameliorates Cognition and Molecular Detrimental Changes after Chronic Mild Stress in SAMP8 Mice.
Puigoriol-Illamola D, Companys-Alemany J, McGuire K, Homer NZM, Leiva R, Vázquez S, Mole DJ, Griñán-Ferré C, Pallàs M. Puigoriol-Illamola D, et al. Pharmaceuticals (Basel). 2021 Oct 13;14(10):1040. doi: 10.3390/ph14101040. Pharmaceuticals (Basel). 2021. PMID: 34681264 Free PMC article.
References
- Rountree M. R. & Selker E. U. DNA methylation and the formation of heterochromatin in Neurospora crassa. Heredity (Edinb.) 105, 38–44 (2010). - PubMed
- Jones P. A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492 (2012). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases