Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity - PubMed (original) (raw)
doi: 10.1038/ncomms12632.
Seung-Oe Lim 1, Weiya Xia 1, Heng-Huan Lee 1, Li-Chuan Chan 1 2, Chu-Wei Kuo 3 4, Kay-Hooi Khoo 4, Shih-Shin Chang 1 2, Jong-Ho Cha 1 5, Taewan Kim 1, Jennifer L Hsu 1 6 7, Yun Wu 8, Jung-Mao Hsu 1, Hirohito Yamaguchi 1, Qingqing Ding 1, Yan Wang 1, Jun Yao 1, Cheng-Chung Lee 3, Hsing-Ju Wu 6, Aysegul A Sahin 8, James P Allison 9, Dihua Yu 1 2, Gabriel N Hortobagyi 10, Mien-Chie Hung 1 2 6 7
Affiliations
- PMID: 27572267
- PMCID: PMC5013604
- DOI: 10.1038/ncomms12632
Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity
Chia-Wei Li et al. Nat Commun. 2016.
Abstract
Extracellular interaction between programmed death ligand-1 (PD-L1) and programmed cell death protein-1 (PD-1) leads to tumour-associated immune escape. Here we show that the immunosuppression activity of PD-L1 is stringently modulated by ubiquitination and N-glycosylation. We show that glycogen synthase kinase 3β (GSK3β) interacts with PD-L1 and induces phosphorylation-dependent proteasome degradation of PD-L1 by β-TrCP. In-depth analysis of PD-L1 N192, N200 and N219 glycosylation suggests that glycosylation antagonizes GSK3β binding. In this regard, only non-glycosylated PD-L1 forms a complex with GSK3β and β-TrCP. We also demonstrate that epidermal growth factor (EGF) stabilizes PD-L1 via GSK3β inactivation in basal-like breast cancer. Inhibition of EGF signalling by gefitinib destabilizes PD-L1, enhances antitumour T-cell immunity and therapeutic efficacy of PD-1 blockade in syngeneic mouse models. Together, our results link ubiquitination and glycosylation pathways to the stringent regulation of PD-L1, which could lead to potential therapeutic strategies to enhance cancer immune therapy efficacy.
Conflict of interest statement
M.-C.H. received sponsored research agreement from STCube Pharmaceuticals Inc. through MD Anderson Cancer Center. C.-W.L., S.-O.L. and M.-C.H. are inventors on patent applications under review: Dual function antibodies specific to glycosylated PD-L1 and methods of use thereof, 2016, No. 62/314,652. Combination treatments directed toward programmed death ligand-1 (PD-LI) positive cancers, 2016, No. 62/316,178. Antibodies specific to glycosylated PD-L1 and methods of use thereof, 2016, No. PCT/US16/24691. The remaining authors declare no competing financial interests.
Figures
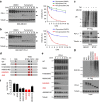
Figure 1. PD-L1 is glycosylated in cancer cells.
(a) Expression of PD-L1 protein in primary breast cancer patient samples. Western blot analysis of PD-L1 in representative breast cancer patient samples. (b) Western blot analysis of PD-L1 in four breast cancer, four melanoma and three lung and three colon cancer cells. (c) Western blot analysis of PD-L1 expression in shCTRL, two independent shPD-L1 stable clones and reconstitution of PD-L1 restored expression in the shPD-L1#5 clone of BT549 cells (left). Western blot analysis of PD-L1 expression in CRISPR/Cas9-mediated PD-L1 knockout BT549 cells (right panel). (d) Glycosylation pattern of PD-L1 protein in BT549 and MDA-MB-231 cells. Cell lysates were treated with PNGase F and analysed by western blot analysis. Black circle, glycosylated PD-L1; arrowhead, non-glycosylated PD-L1. (e) Glycosylation pattern of PD-L1-GFP, HA-PD-L1 and PD-L1-Flag proteins. Cell lysates were treated with PNGase F and Endo H and analysed by western blot analysis. (f) Schematic diagram of PD-L1 protein. Full-length PD-L1 was separated into extracellular domain (ECD) and intracellular domain (ICD). SP, signal peptide; TM, transmembrane domain. Four putative NXT motifs in the ECD domain are labelled in red. The numbers indicate amino-acid positions. (g) LC-MS/MS-based identification of _N_-glycopeptides. LC-MS/MS-based identification of _N_-glycopeptides corresponding to one of the four _N_-glycosylation sites, N35. The LC-MS profiles (top) are shown as spectra averaged over a period of elution time (as labelled in figures) when a representative subset of glycoforms were detected. For each _N_-glycosylation site, one representive HCD MS2 spectrum (bottom) is shown to exemplify its identification based on detection of y1 ion (tryptic peptide backbone carrying the GlcNAc attached to the _N_-glycosylated Asn), along with the b and y ions defining its peptide sequence. The cartoon symbols used for the glycans (see inset) conform to the standard representation recommended by the Consortium for Functional Glycomics. (h) Western blot analysis of the protein expression pattern of PD-L1 WT and its NQ mutants. Non-glycosylated form in lane 14 indicates PD-L1 WT with overnight treatment with TM. Black circle, glycosylated PD-L1; arrowhead, non-glycosylated PD-L1.
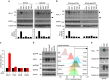
Figure 2. Glycosylation of PD-L1 stabilizes PD-L1 protein.
(a,b) Western blot analysis of PD-L1 protein in PD-L1-Flag expressing MDA-MB-231 cells (a) and HEK 293T cells (b). Cells were treated with 20 μM cycloheximide (CHX) at indicated intervals and analysed by western blot analysis. The intensity of PD-L1 protein was quantified using a densitometer. (c) Inhibition of PD-L1 glycosylation enhances ubiquitination. Breast cancer cells with TM and/or MG132 treatment were subjected to PD-L1 immunoprecipitation (IP) and western blot analyses with anti-K48 ubiquitin. (d) Schematic diagram of various PD-L1 NQ mutants used in this study. The numbers indicate amino acid positions on the PD-L1. (e) Protein half-life of PD-L1 WT or various NQ mutants expressing in MDA-MB-231 cells. Experiments were performed as described in (a). Quantification of PD-L1 half-life was shown in (f). (g) Ubiquitination of PD-L1 proteins in PD-L1 WT or various NQ mutants expressing MDA-MB-231 cells. PD-L1 proteins were IP with Flag antibody and then immunoblotted with ubiquitin antibody. Approximately 5% of the cell extract from IP was saved as input. Black circle, glycosylated PD-L1; arrowhead, non-glycosylated PD-L1. All error bars are expressed as mean±s.d. of 3 independent experiments.
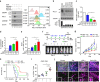
Figure 3. GSK3β binds to and phosphorylates PD-L1 protein in vitro and in vivo.
(a) Schematic diagram of GSK3β phosphorylation and β-TrCP-binding motifs and various mutants of PD-L1 expression constructs. PD-L1 was separated into ECD and ICD. SP, signal peptide; TM, transmembrane domain. The numbers indicate amino-acid positions. (b) In vitro GST pull-down assay of non-glycosylated PD-L1 and GSK3β. (c) Co-immunoprecipitation (co-IP) measuring the interaction of GSK3β and PD-L1 4NQ. Schematic diagram of PD-L1 4NQ deletion or truncation mutants showing on the left. Positions of glycosylation sites were labelled with red colour. The numbers indicate amino-acid positions. (d) Immunocomplex kinase assay measuring PD-L1 phosphorylation by GSK3β. Coomassie blue staining showing equal loading amount of GST-PD-L1. CA, constitutive activation mutant (S9A); KD, kinase dead (K85A) mutant; WT, wild type. (e) Western blot analysis of phosphorylation of PD-L1 protein at T180 and S184 sites by phospho-T180 and -S184 PD-L1 antibodies, respectively. EV, empty vector. (f) Time-lapse microscopy image (at 12 h) showing the dynamic interaction between PD-L1 and PD-1 at the last time point. The kinetic graph showed the quantitative binding of green fluorescent labelled PD-1/Fc protein on PD-L1 WT, 3SA or 4NQ expressing BT549 cells at every hour time point (right). Scale bar, 100 μm. (g) T-cell-meditated tumour cell-killing assay in PD-L1 WT or 3SA-expressing BT549 cells. Representative phase, red fluorescent (nuclear-restricted RFP), and/or green fluorescent (Caspase 3/7 substrate)-merged images of PD-L1 WT- or PD-L1 3SA-expressing cells and activated T-cell co-cultures at 96 h. Green fluorescent cell was counted as dead cell. The quantitative ratio of dead cells showed in bar graph (right). Scale bar, 100 μm. (h) The tumour growth of mouse PD-L1 WT- or PD-L1 3SA-expressing 4T1 cells in BALB/c mice. Quantification of tumour volume is shown on the right and representative images of tumours are shown on the left. _n_=7 mice per group. Con, vector control; WT, PD-L1 WT; 3SA, PD-L1 3SA. *P<0.05 is statistically significant as shown by Student's _t_-test. All error bars are expressed as mean±s.d. of three independent experiments.
Figure 4. EGF signalling induces PD-L1 glycosylation.
(a) Western blot analysis of PD-L1 expression in BT549 and MB-468 cells treated with 25 ng ml−1 EGF, 25 ng ml−1 insulin-like growth factor-1, 10 ng ml−1 hepatocyte growth factor, 25 ng ml−1 fibroblast growth factor and 100 nM TGFβ for overnight. shCTRL, control shRNA. (b) Western blot analysis of PD-L1 expression in BT549-shCTRL and BT549-shEGFR cells. (c) Quantification of western blot results from Supplementary Fig. 8a. Cells were in a serum-free culture medium for overnight and then treated with EGF. The intensity of PD-L1 protein was quantified using a densitometer. (d) Western blot analysis of PD-L1 expression upon different agonist treatments for overnight. Cell surface analysis of PD-L1 protein using flow cytometer is shown in the right. (e) Flag-tagged PD-L1 was stably expressed in BT549 cells. Western blot analysis showing exogenous PD-L1 expression under EGF treatment.
Figure 5. Inhibition of EGFR sensitizes the PD-1 blockade therapy in syngeneic mouse model.
(a) Cells were treated with TKIs for 2 h before EGF stimulation. Cell surface analysis of PD-L1 protein using flow cytometer was shown in the right. (b) Western blot analysis of PD-L1 protein in the cells treated with several indicated inhibitors. PD-L1 WT-expressing BT549 cells were treated with 1 μg ml−1 TM, 1 μM gefitinib, 1 μM erlotinib, 1 μM lapatinib and AG1478. (c) PD-L1 and PD-1 interaction in PD-L1-expressing BT549 cells. (d) Soluble IL-2 levels in PD-L1-expressing BT549 cells treated with gefitinib and/or anti-PD-1 antibody. (e) T-cell-meditated killing of PD-L1-expressing BT549 cells treated with gefitinib and/or anti-PD-1 antibody. (f) The tumour growth of 4T1-Luc cells in BALB/c mice following treatment with gefitinib and/or anti-PD-1 antibody. Treatment protocol is summarized (top). Tumour growth of 4T1-Luc cells was shown in vivo by bioluminescence imaging using IVIS100 (bottom). (g) The tumour growth of 4T1 cells in gefitinib- and/or anti-PD-1 antibody-treated BALB/c mice. Tumours were measured at the indicated time points and dissected at end point. Quantification of tumour volume is shown on the right and representative images of tumours are shown on the left. _n_=9 mice per group. (h) Survival of mice bearing syngeneic 4T1-Luc-derived tumour following treatment with gefitinib and/or anti-PD-1 antibody. Significance was determined by log-rank test. *P<0.05; _n_=10 mice per group. (i) Intracellular cytokine stain of IFNγ and CD8 in CD3+ T-cell populations from the isolated tumour-infiltrating lymphocytes. (j) Immunofluorescence staining of the protein expression pattern of PD-L1, CD8 and granzyme B (GB) in 4T1 tumour mass. *P<0.05 is statistically significant as shown by Student's _t-_test. All error bars are expressed as mean±s.d. of three independent experiments. Gef, gefitinib.
Similar articles
- Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance.
Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, Tan Y, Ci Y, Wu F, Dai X, Guo J, Huang YH, Fan C, Ren S, Sun Y, Freeman GJ, Sicinski P, Wei W. Zhang J, et al. Nature. 2018 Jan 4;553(7686):91-95. doi: 10.1038/nature25015. Epub 2017 Nov 16. Nature. 2018. PMID: 29160310 Free PMC article. - FAT4 overexpression promotes antitumor immunity by regulating the β-catenin/STT3/PD-L1 axis in cervical cancer.
Wang D, Wu S, He J, Sun L, Zhu H, Zhang Y, Liu S, Duan X, Wang Y, Xu T. Wang D, et al. J Exp Clin Cancer Res. 2023 Sep 1;42(1):222. doi: 10.1186/s13046-023-02758-2. J Exp Clin Cancer Res. 2023. PMID: 37658376 Free PMC article. - Metformin Suppresses Both PD-L1 Expression in Cancer Cells and Cancer-Induced PD-1 Expression in Immune Cells to Promote Antitumor Immunity.
Park SH, Lee J, Yun HJ, Kim SH, Lee JH. Park SH, et al. Ann Lab Med. 2024 Sep 1;44(5):426-436. doi: 10.3343/alm.2023.0443. Epub 2024 Mar 26. Ann Lab Med. 2024. PMID: 38529546 Free PMC article. - Posttranslational Modifications of PD-L1 and Their Applications in Cancer Therapy.
Hsu JM, Li CW, Lai YJ, Hung MC. Hsu JM, et al. Cancer Res. 2018 Nov 15;78(22):6349-6353. doi: 10.1158/0008-5472.CAN-18-1892. Cancer Res. 2018. PMID: 30442814 Free PMC article. Review. - PD-1/PD-L1 Pathway in Breast Cancer.
Schütz F, Stefanovic S, Mayer L, von Au A, Domschke C, Sohn C. Schütz F, et al. Oncol Res Treat. 2017;40(5):294-297. doi: 10.1159/000464353. Epub 2017 Mar 27. Oncol Res Treat. 2017. PMID: 28346916 Review.
Cited by
- PD-L1 recruits phospholipase C and enhances tumorigenicity of lung tumors harboring mutant forms of EGFR.
Ghosh S, Nataraj NB, Noronha A, Patkar S, Sekar A, Mukherjee S, Winograd-Katz S, Kramarski L, Verma A, Lindzen M, Garcia DD, Green J, Eisenberg G, Gil-Henn H, Basu A, Lender Y, Weiss S, Oren M, Lotem M, Geiger B, Ruppin E, Yarden Y. Ghosh S, et al. Cell Rep. 2021 May 25;35(8):109181. doi: 10.1016/j.celrep.2021.109181. Cell Rep. 2021. PMID: 34038737 Free PMC article. - Upregulation of PD-L1 in Senescence and Aging.
Onorati A, Havas AP, Lin B, Rajagopal J, Sen P, Adams PD, Dou Z. Onorati A, et al. Mol Cell Biol. 2022 Oct 20;42(10):e0017122. doi: 10.1128/mcb.00171-22. Epub 2022 Sep 26. Mol Cell Biol. 2022. PMID: 36154662 Free PMC article. - Exploring the Role of PD-1 in the Autoimmune Response: Insights into Its Implication in Systemic Lupus Erythematosus.
Sagrero-Fabela N, Chávez-Mireles R, Salazar-Camarena DC, Palafox-Sánchez CA. Sagrero-Fabela N, et al. Int J Mol Sci. 2024 Jul 15;25(14):7726. doi: 10.3390/ijms25147726. Int J Mol Sci. 2024. PMID: 39062968 Free PMC article. Review. - Removal of N-Linked Glycosylation Enhances PD-L1 Detection in Colon Cancer: Validation Research Based on Immunohistochemistry Analysis.
Xu J, Yang X, Mao Y, Mei J, Wang H, Ding J, Hua D. Xu J, et al. Technol Cancer Res Treat. 2021 Jan-Dec;20:15330338211019442. doi: 10.1177/15330338211019442. Technol Cancer Res Treat. 2021. PMID: 34060360 Free PMC article. - A functional role for glycosylated B7-H5/VISTA immune checkpoint protein in metastatic clear cell renal cell carcinoma.
Emaldi M, Alamillo-Maeso P, Rey-Iborra E, Mosteiro L, Lecumberri D, Pulido R, López JI, Nunes-Xavier CE. Emaldi M, et al. iScience. 2024 Jul 25;27(9):110587. doi: 10.1016/j.isci.2024.110587. eCollection 2024 Sep 20. iScience. 2024. PMID: 39262813 Free PMC article.
References
- Spranger S., Bao R. & Gajewski T. F. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 523, 231–235 (2015). - PubMed
- Dong H. et al.. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat. Med. 8, 793–800 (2002). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials