Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells - PubMed (original) (raw)
Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells
Gianluigi Lichinchi et al. Nat Microbiol. 2016.
Abstract
N(6)-methyladenosine (m(6)A) is the most prevalent internal modification of eukaryotic mRNA. Very little is known of the function of m(6)A in the immune system or its role in host-pathogen interactions. Here, we investigate the topology, dynamics and bidirectional influences of the viral-host RNA methylomes during HIV-1 infection of human CD4 T cells. We show that viral infection triggers a massive increase in m(6)A in both host and viral mRNAs. In HIV-1 mRNA, we identified 14 methylation peaks in coding and noncoding regions, splicing junctions and splicing regulatory sequences. We also identified a set of 56 human gene transcripts that were uniquely methylated in HIV-1-infected T cells and were enriched for functions in viral gene expression. The functional relevance of m(6)A for viral replication was demonstrated by silencing of the m(6)A writer or the eraser enzymes, which decreased or increased HIV-1 replication, respectively. Furthermore, methylation of two conserved adenosines in the stem loop II region of HIV-1 Rev response element (RRE) RNA enhanced binding of HIV-1 Rev protein to the RRE in vivo and influenced nuclear export of RNA. Our results identify a new mechanism for the control of HIV-1 replication and its interaction with the host immune system.
Figures
Figure 1.. m6A RNA methylation in T cells is promoted by HIV-1 infection and modulates viral replication.
a, Analysis of RNA from HIV-1-infected T cells. Total RNA extracted from uninfected (Ctrl) or HIV-1-infected (HIV) MT4 cells 3 days post-infection was analysed by ethidium bromide (EtBr) staining (left), anti-m6A immunoblotting (middle), or northern blotting for HIV-1 RNA (right). Arrow indicates the predicted size of full-length HIV-1 RNA. The data is a representative of seven independent experiments. b, LC-MS/MS quantification of m6A content in poly(A) enriched RNA fractions. Results are the mean ± s.e.m. of five independent experiments. P values were calculated by unpaired two-tailed Student’s _t_-test. c, d, HIV-1 replication is reduced by METTL3 and METTL14 silencing and enhanced by AlkBH5 silencing. c, RT-qPCR analysis of gp120 expression in MT4 cells transduced for 3 days with nontargeting control (NTC), METTL3, METTL14, or AlkBH5 shRNA. Cells were then infected with HIV-1 LAI virus, and cells were analysed 3 days later. d, Western blot analysis of HIV-1 p24 intracellular protein expression in cells described in c. Results are the mean ± s.e.m. of four independent experiments. P values were calculated by unpaired two-tailed Student’s _t_-test.
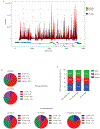
Figure 2.. RNA methylation profiles of HIV-1 and HIV-1-infected T cells.
a, Methylation peaks in the full-length HIV-1 RNA genome. Viral gene annotation is shown below. Poly(A) RNA from uninfected and HIV-1-infected MT4 cells was fragmented, subjected to RNA-seq and MeRIP-seq, and analysed as described previously. The grey shading corresponds to the total RNA-seq mapping distribution (MeRIP input), and the red signal corresponds to the MeRIP-seq mapping distribution. On the x-axis, the dark blue boxes and numbers indicate the m6A peaks called by MeRIPPeR analysis and referred to in the text. b, Topology of m6A in the cellular and viral transcriptomes. Pie charts showing the distribution of total RNA sequence (upper) and m6A peaks (middle) in four regions (5′UTR, CDS, 3′UTR, and intronic) of transcripts from uninfected and HIV-1-infected MT4 cells. m6A peak distribution in HIV-1-specific RNA is also shown (bottom). c, Frequency of TGAC, RRACH, and MGACK sequence motifs within m6A peaks in cellular RNAs from control and HIV-1-infected MT4 cells (Ctrl_cellular RNAs and HIV_cellular RNAs, respectively) and HIV-1 LAI RNA (HIV_HIV RNA).
Figure 3.. m6A methylation modulates the interaction between Rev protein and RRE RNA.
a, Secondary structure of the hairpin loop IIB of HIV-1 RRE. The two highlighted adenosines represent the methylation sites identified in peak 11 on Figure 2a. Nucleotide numbering is based on the pLAI2 proviral vector sequence, and the mutation frequency of the adenosines in 2,501 HIV-1 sequences is shown in parentheses. b, RRE methylation enhances Rev interactions. 293T cells were depleted of METTL3, METTL14, or AlkBH5, and transfected pLAI2 vector. RIP-qPCR experiments were performed by IP with control IgG or anti-Rev antibody followed by qPCR to analyse RRE RNA. Results are the mean ± s.e.m. of 3 independent experiments and are expressed as the fold enrichment of RRE RNA in anti-Rev versus control IgG IPs. P values were calculated by unpaired two-tailed Student’s _t_-test. c, RRE A7877 and A7883 are methylated in vivo. Primers were designed to probe for m6A modification at these two sites in RRE. AMV and Tth primer extensions for A7877 and A7883 were performed on 1 µg poly(A) enriched RNA from HIV-1 infected MT4 cells. Extension for 28S A4189 and 28S A4190 were performed on 10 µg total RNA and served as negative and positive methylation sites, respectively.
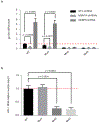
Figure 4.. HIV-1 replication and RNA nuclear export are perturbed by methylation of A7883 in the RRE bulge region.
a, WT or LAI HIV-1 mutant constructs, U7871C + A7877G (Mut1), A7883G (Mut2), and the combination of both (Mut3), were assessed for replication in 293T cells under METTL3/METTL14 or AlkBH5 knock-down conditions. Results are the mean ± s.e.m. of three independent experimentsand P values were calculated by unpaired two-tailed Student’s _t_-test. b, Nuclear export of viral RNA was analysed by cytoplasmic and nuclear fractionation in 293T cells transfected with WT or mutant LAI constructs described in a. Viral RNA distribution in the two compartments is relative to the viral RNA content from total cellular RNA. Results are the mean ± s.e.m. of three independent experiments. P values were calculated by unpaired two-tailed Student’s _t_-test.
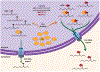
Figure 5.
Proposed model for modulation of the m6A modification in RRE RNA and its effect on HIV-1 replication. See text for details. For simplicity, only monomeric Rev–RRE interactions are shown.
Comment in
- Viral cell biology: HIV RNA gets methylated.
Ciuffi A. Ciuffi A. Nat Microbiol. 2016 Mar 29;1:16037. doi: 10.1038/nmicrobiol.2016.37. Nat Microbiol. 2016. PMID: 27572449 No abstract available.
Similar articles
- N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression.
Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L. Tirumuru N, et al. Elife. 2016 Jul 2;5:e15528. doi: 10.7554/eLife.15528. Elife. 2016. PMID: 27371828 Free PMC article. - Method for the Enrichment of N6-Methyladenosine-Modified Cellular and HIV-1 RNA.
Mishra T, Phillips S, Wu L. Mishra T, et al. Methods Mol Biol. 2024;2807:195-208. doi: 10.1007/978-1-0716-3862-0_14. Methods Mol Biol. 2024. PMID: 38743230 Free PMC article. - Evolution of the HIV-1 Rev Response Element during Natural Infection Reveals Nucleotide Changes That Correlate with Altered Structure and Increased Activity over Time.
Sherpa C, Jackson PEH, Gray LR, Anastos K, Le Grice SFJ, Hammarskjold ML, Rekosh D. Sherpa C, et al. J Virol. 2019 May 15;93(11):e02102-18. doi: 10.1128/JVI.02102-18. Print 2019 Jun 1. J Virol. 2019. PMID: 30867301 Free PMC article. - Viral Epitranscriptomics.
Kennedy EM, Courtney DG, Tsai K, Cullen BR. Kennedy EM, et al. J Virol. 2017 Apr 13;91(9):e02263-16. doi: 10.1128/JVI.02263-16. Print 2017 May 1. J Virol. 2017. PMID: 28250115 Free PMC article. Review. - N6-methyladenosine modification in mRNA: machinery, function and implications for health and diseases.
Maity A, Das B. Maity A, et al. FEBS J. 2016 May;283(9):1607-30. doi: 10.1111/febs.13614. Epub 2015 Dec 31. FEBS J. 2016. PMID: 26645578 Review.
Cited by
- Sequencing methods and functional decoding of mRNA modifications.
Li K, Peng J, Yi C. Li K, et al. Fundam Res. 2023 Jun 1;3(5):738-748. doi: 10.1016/j.fmre.2023.05.010. eCollection 2023 Sep. Fundam Res. 2023. PMID: 38933299 Free PMC article. Review. - Post-transcriptional gene regulation by mRNA modifications.
Zhao BS, Roundtree IA, He C. Zhao BS, et al. Nat Rev Mol Cell Biol. 2017 Jan;18(1):31-42. doi: 10.1038/nrm.2016.132. Epub 2016 Nov 3. Nat Rev Mol Cell Biol. 2017. PMID: 27808276 Free PMC article. Review. - Integrative network analysis identifies cell-specific trans regulators of m6A.
An S, Huang W, Huang X, Cun Y, Cheng W, Sun X, Ren Z, Chen Y, Chen W, Wang J. An S, et al. Nucleic Acids Res. 2020 Feb 28;48(4):1715-1729. doi: 10.1093/nar/gkz1206. Nucleic Acids Res. 2020. PMID: 31912146 Free PMC article. - Infection Meets Inflammation: N6-Methyladenosine, an Internal Messenger RNA Modification as a Tool for Pharmacological Regulation of Host-Pathogen Interactions.
Leseva MN, Buttari B, Saso L, Dimitrova PA. Leseva MN, et al. Biomolecules. 2023 Jun 29;13(7):1060. doi: 10.3390/biom13071060. Biomolecules. 2023. PMID: 37509095 Free PMC article. Review. - HibeRNAtion: HIV-1 RNA Metabolism and Viral Latency.
Crespo R, Rao S, Mahmoudi T. Crespo R, et al. Front Cell Infect Microbiol. 2022 Jun 14;12:855092. doi: 10.3389/fcimb.2022.855092. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35774399 Free PMC article. Review.
References
- Desrosiers RC, Friderici KH & Rottman FM Characterization of Novikoff hepatoma mRNA methylation and heterogeneity in the methylated 5' terminus. Biochemistry 14, 4367–4374 (1975). - PubMed
- Perry RP, Kelley DE, Friderici K & Rottman F The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5' terminus. Cell 4, 387–394 (1975). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials